Difference between revisions of "20.109(S19):Module 1"
Noreen Lyell (Talk | contribs) (Created page with "<div style="padding: 10px; width: 820px; border: 5px solid #0404B4;"> {{Template:20.109(S19)}} =<center>Module 1</center>= '''Lecturer:''' [http://be.mit.edu/directory/ang...") |
(→References) |
||
| (8 intermediate revisions by 2 users not shown) | |||
| Line 6: | Line 6: | ||
'''Lecturer:''' [http://be.mit.edu/directory/angela-koehler Angela Koehler]<br> | '''Lecturer:''' [http://be.mit.edu/directory/angela-koehler Angela Koehler]<br> | ||
| − | '''Instructors:''' [http://be.mit.edu/directory/noreen-lyell Noreen Lyell] and [http://be.mit.edu/directory/leslie-mcclain Leslie McClain] | + | '''Instructors:''' [http://be.mit.edu/directory/noreen-lyell Noreen Lyell] and [http://be.mit.edu/directory/leslie-mcclain Leslie McClain]<br> |
| − | + | ||
'''TAs:''' Catherine Henry and Michaela Gold <br> | '''TAs:''' Catherine Henry and Michaela Gold <br> | ||
'''Lab manager:''' Hsinhwa Lee <br> | '''Lab manager:''' Hsinhwa Lee <br> | ||
| Line 20: | Line 19: | ||
This module has been developed thanks to the generous time and thoughtful efforts of several Koehler Laboratory members, in particular Rob Wilson, Dr. Becky Leifer, and Shelby Doyle. | This module has been developed thanks to the generous time and thoughtful efforts of several Koehler Laboratory members, in particular Rob Wilson, Dr. Becky Leifer, and Shelby Doyle. | ||
| − | [[Image: | + | [[Image:Sp19_Mod1_overview_schematic.jpg|thumb|750px|center|]] |
<br style="clear:both" /> | <br style="clear:both" /> | ||
| Line 26: | Line 25: | ||
==Lab links: day by day== | ==Lab links: day by day== | ||
M1D1: [[20.109(S19):In silico cloning and confirmation digest of protein expression vector(Day1) |In silico cloning and confirmation digest of protein expression vector]]<br> | M1D1: [[20.109(S19):In silico cloning and confirmation digest of protein expression vector(Day1) |In silico cloning and confirmation digest of protein expression vector]]<br> | ||
| − | M1D2: [[20.109(S19): | + | M1D2: [[20.109(S19):Purify protein for secondary assays(Day2) |Purify protein for secondary assays]]<br> |
| − | M1D3: [[20.109( | + | M1D3: [[20.109(S18):Analyze small microarray data (Day3) | Analyze small molecule microarray (SMM) data]]<br> |
M1D4: [[20.109(S19):Evaluate protein purity and concentration (Day4) |Evaluate protein purity and concentration]]<br> | M1D4: [[20.109(S19):Evaluate protein purity and concentration (Day4) |Evaluate protein purity and concentration]]<br> | ||
M1D5: [[20.109(S19):Test protein activity using peptidyl-prolyl cis-trans isomerase assay (Day5) |Test protein activity using peptidyl-prolyl cis-trans isomerase assay]]<br> | M1D5: [[20.109(S19):Test protein activity using peptidyl-prolyl cis-trans isomerase assay (Day5) |Test protein activity using peptidyl-prolyl cis-trans isomerase assay]]<br> | ||
| Line 34: | Line 33: | ||
==Assignments== | ==Assignments== | ||
| − | [[20.109(S19): | + | [[20.109(S19): Data Summary|Data summary]]<br> |
| − | [[20.109(S19): | + | [[20.109(S19): Mini-presentation| Mini-presentation]] <br> |
==References== | ==References== | ||
| Line 42: | Line 41: | ||
*[[Media:Sp17 M1 reference ChemBiol.pdf| Recent discoveries and applications involving small-molecule microarrays.]] ''Chemical Biology.'' 18:21-28. | *[[Media:Sp17 M1 reference ChemBiol.pdf| Recent discoveries and applications involving small-molecule microarrays.]] ''Chemical Biology.'' 18:21-28. | ||
| + | |||
| + | *[[Media:Sp19 M1 reference Cell.pdf| Ligand and Target Discovery by Fragment-Based Screening in Human Cells.]] ''Cell.'' 168, 527–541. | ||
| + | |||
| + | *[[Media:Sp19 M1 reference JVisExp.pdf| Determination of Protein-ligand Interactions Using Differential Scanning Fluorimetry.]] ''J Vis Exp.'' 91:51809. | ||
==Notes for teaching faculty== | ==Notes for teaching faculty== | ||
[[20.109(S19): Prep notes for M1| Prep notes for M1]] | [[20.109(S19): Prep notes for M1| Prep notes for M1]] | ||
Latest revision as of 16:35, 24 February 2019
Contents
Module 1
Lecturer: Angela Koehler
Instructors: Noreen Lyell and Leslie McClain
TAs: Catherine Henry and Michaela Gold
Lab manager: Hsinhwa Lee
Overview
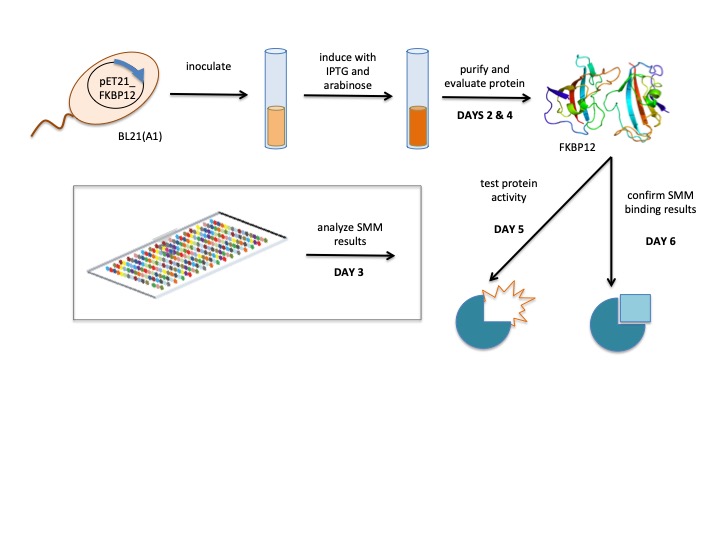
Chemical probes, or ligands, are important research tools used to explore cellular processes and therapeutic targets. The use of high-throughput and unbiased strategies to identify small molecules that bind specific biomolecules, such as proteins, can provide insight on the structure or function of targets. Additionally, a small-molecule screen can identify new chemical probes for target proteins of interest.
The small-molecule microarray (SMM) is a high-throughput method that enables the detection of protein-ligand binding. Briefly, ligands are 'printed' onto a slide and incubated with purifed protein. Unbound protein is washed from the slide and bound protein is detected using a tag on the protein of interest. Because the location of every ligand on the slide is known, the detection of protein indicates that it is bound to the ligand at that location.
In your experiment, you will use SMM data collected by students from the Sp17 semester to identify ligands that bind to FKBP12, a folding chaperone for proteins that contain proline residues in eukaryotes. To expand on the research completed by the previous 109ers, you will confirm that the identified ligands bind FKBP12 using secondary assays.
This module has been developed thanks to the generous time and thoughtful efforts of several Koehler Laboratory members, in particular Rob Wilson, Dr. Becky Leifer, and Shelby Doyle.
Lab links: day by day
M1D1: In silico cloning and confirmation digest of protein expression vector
M1D2: Purify protein for secondary assays
M1D3: Analyze small molecule microarray (SMM) data
M1D4: Evaluate protein purity and concentration
M1D5: Test protein activity using peptidyl-prolyl cis-trans isomerase assay
M1D6: Confirm ligand binding using differential scanning fluorimetry assay
M1D7: Complete data analysis
Assignments
Data summary
Mini-presentation
References
- A method for the covalent capture and screening of diverse small molecules in a microarray format. Nature Protocols. 1:2344-2352.
- Recent discoveries and applications involving small-molecule microarrays. Chemical Biology. 18:21-28.
- Ligand and Target Discovery by Fragment-Based Screening in Human Cells. Cell. 168, 527–541.
- Determination of Protein-ligand Interactions Using Differential Scanning Fluorimetry. J Vis Exp. 91:51809.