20.109(S19):Test protein activity using peptidyl-prolyl cis-trans isomerase assay (Day5)
Contents
Introduction
Today you will test the activity of FKBP12 using a peptidyl-prolyl isomerase (PPIase) assay. PPIases catalyze cis-trans isomerization reactions that are essential to efficient protein folding in vivo. Specifically, these enzymes isomerize peptide bonds that are N-terminal to proline residues in polypeptide chains. Without PPIases, isomerization would be the rate-limiting step in protein folding.
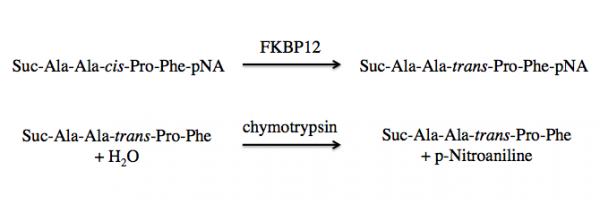
FKBP12 is a class of PPIases and the activity can be measured by quantifying the isomerization and subsequent cleavage of a substrate, suc-AAFP-pNA (also written as Suc-Ala-Ala-Pro-Phe-NA). In this method, FKBP12 catalyzes the isomeration of the cis-Ala-Pro bond to a trans-Ala-Pro bond. Then a second enzyme, chymytrypsin, cleaves the trans form of the peptide. See the reaction schematic below.
When at equilibrium in solution, approximately 88% of the commercially available suc-AAFP-pNA substrate is in the trans from, leaving only a small amount of the peptide for isomerization. Because of this, the suc-AAFP-pNA substrate is prepared in trifluoroethanol (TFE) containing lithium chloride, LiCl. The Li+ ions maintain the substrate in 60% cis form.
Upon cleavage by chymotrypsin, the release of p-nitroaniline results in a yellow color in alkaline conditions, which absorbance can be measured at 405 nm. The rate of ΔA405 nm (change in absorbance at 405 nm) in the presence versus absence of FKBP12 is used to calculate the activity.
Protocols
Part 1: Prepare samples for PPIase assay
For your planning, each team will setup triplicate reactions for 4 different conditions. The FKBP12 protein will not be added until the end, and the suc-AAFP-pNA substrate will be loaded by the teaching faculty just prior to measurement. The subsequent steps will help you create the proper conditions, which are outlined in the below table (a 'Y' indicates the component is included in the condition / reaction).
| Condition #1 'NO substrate' Control |
Condition #2 'NO chymotrypsin' Control |
Condition #3 'NO protein' Control |
Condition #4 Experimental | |
|---|---|---|---|---|
| Assay buffer | Y | Y | Y | Y |
| Chymotrypsin | Y | Y | Y | |
| FKBP12 protein | Y | Y | Y | |
| suc-AAFP-pNA (substrate) | Y | Y | Y |
- Calculate the volumes of each component you will need for each of the reaction conditions using the following information:
- The total volume of each reaction is 200 μL.
- The reaction buffer consists of 200 mM Tris-HCl, pH = 8 (stock concentration is 1 M) and 0.2% v/v DMSO (stock concentration is 100%).
- A volume of 20 μL of substrate is added to each reaction.
- In the NO substrate condition, reaction buffer is used to account for the difference in volume.
- The final concentration of chymotrypsin is 20 nM (stock concentration is 20 μM) in each reaction.
- In the NO chymotrypsin condition, reaction buffer is used to account for the difference in volume.
- A total of 1 μg of protein is added to each reaction. The volume is based on the concentration you calculated during the previous laboratory session.
- In the NO protein condition, 1X PBS is used to account for the difference in volume.
- Use sterile H2O as the diluent.
- Use a table or chart to write the volumes of each component needed for each reaction condition; then confirm your numbers with the teaching faculty.
- Clearly label four 1.5 mL microcentrifuge tubes, one for each condition.
- Prepare a 'master mix' for each of the four conditions.
- You will test each condition in triplicate, but need to prepare enough master mix for 3.25 reactions to account for possible pipetting errors.
- IMPORTANT NOTE: do NOT add protein until instructed to do so by the teaching faculty!
- Keep your master mixes on ice.
- The teaching faculty will add the substrate immediately prior to data collection.
- Alert the teaching faculty when your master mixes are prepared.
- When told to do so, add the appropriate volume of protein solution to the appropriate master mixes and mix well.
- You will transfer your master mixes into a 96-well plate on the front laboratory bench.
- The teaching faculty will tell you which wells to use as multiple teams will add their samples to the same plate.
- Transfer 180 μL of each master mix into 3 wells according to the plate map.
- Be mindful that you are adding your samples to the correct wells!
After all teams have loaded their samples, the teaching faculty will add 20 μL of the suc-AAFP-pNA substrate (5 mM in 3 mM TFE containing 460 mM LiCl) to the samples and immediately load the plate onto the plate reader. The substrate must be added at the end to ensure that the reaction does not take place before its absorbance is being monitored by the plate reader. The plate reader will record the A405 nm values at 25 °C for each sample every minute for 30 minutes (a total of 31 readings including t = 0).
Part 2: Calculate activity of FKBP12
The data for your PPIase experiment will be provided as an excel spreadsheet that includes the A405 nm values for each well at every timepoint. You are responsible for the data analysis of only your samples, not for the entire plate.
- Average the triplicate wells for each condition you tested at every timepoint.
- Plot the averaged absorbance values (y-axis) over time (x-axis) for each condition.
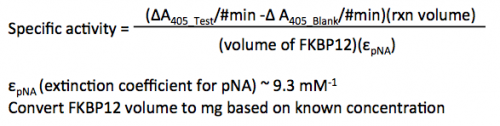
- Use the t = 0 and t = 30 to calculate the activity of FKBP12 condition tested.
- TEST = Condition 4, and BLANK = Condition 3.
- Be sure to use the appropriate value for minutes given the t values.
- The calculations are completed for each condition separately.
- Post an Excel spreadsheet containing the conditions you tested, plots, and the nmoles/min/mg protein value calculated for each condition to Class data page.
Reagents
- Chymotrypsin (Sigma)
- suc-AAFP-pNA (Sigma
- 1 M Tris-HCl, ph=8 (Boston BioProducts)
- DMSO
Next day: Confirm ligand binding using differential scanning fluorimetry assay