20.109(S19):Evaluate protein purity and concentration (Day4)
Contents
Introduction
Electrophoresis is a technique that separates large molecules by size using an applied electrical field and a sieving matrix. DNA, RNA and proteins are the molecules most often studied with this technique; agarose and acrylamide gels are the two most common sieves. The molecules to be separated enter the matrix through a well at one end and are pulled through the matrix when a current is applied across it. The larger molecules get entwined in the matrix and are stalled; the smaller molecules wind through the matrix more easily and travel farther away from the well. The distance a nucleic acid or amino acid fragment travels is inversely proportional to the log of its length. Over time fragments of similar length accumulate into “bands” in the gel.
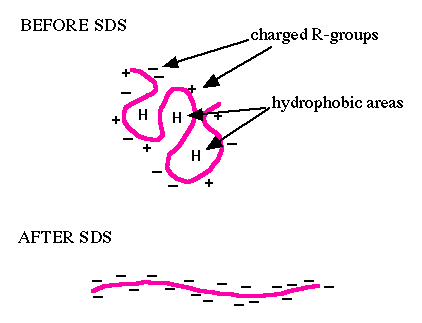
You will use sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to evaluate your purified protein. SDS is an ionic surfactant (or detergent), which denatures the proteins and coats them with a negative charge. Since denatured proteins are linear, they will move through the gel at a speed inversely proportional to their molecular weight, just like DNA on agarose gels. (Non-denatured proteins run according to their molecular weight, shape, and charge.) You will run a reference ladder containing proteins of known molecular weight and amount to determine the size and concentration of your purified FKBP12 protein. When running uninduced and induced samples side-by-side, you should see a protein band at the expected molecular weight for FKBP12 (~14 kDa + 3 kDa from the His-tag = 17 kDa), which may be very faint or non-existent in the uninduced control sample, but bright and thick in the induced sample. To visualize all the proteins released by the bacteria, you will stain the gels with Coomassie Brilliant Blue (actually, a variant called BioSafe Coomassie). Because this is a non-specific stain for all proteins, it will provide information concerning the purity of your protein sample.To measure the concentration of your purified protein, you will use the BCA Protein Assay Reagent Kit. This kit enables colorimetric detection and quantification of the total protein within a sample. The ability to measure protein concentration is based on the detection of Cu1+ by the detection reagent, bicinchoninic acid (BCA). The Cu1+ is formed when Cu2+ is reduced by protein in an alkaline environment. Through this reduction reaction, a purple product is formed by the chelation of BCA and Cu1+ at a 2:1 ratio. This water-soluble complex has a strong absorbance at 562 nm. Because absorbance and protein concentration have a linear relationship, it is possible to compare the absorbance of an unknown protein sample to a standard curve, generated with samples of known protein concentrations, and calculate the concentration of protein in the experimental sample.
Protocols
Part 1: Desalt purified FKBP12 solution
Before your purified protein can be used for your experiments you will desalt the preparation with PBS buffer to remove the imidazole, which may interfere with the secondary assays.
- Obtain your purified protein solution and a desalting columnn from the front laboratory bench.
- Remove the bottom of the desalting column and place the column in a 15 mL conical tube with the cap loosely attached.
- Centrifuge for 2 min at 1000 g (2000 rpm) then discard the flowthrough.
- Rinse the desalting column by completing the following steps:
- Add 2.5 mL of 1X PBS.
- Centrifuge for 2 min at 1000 g then discard the flowthrough.
- Complete this process a total of 2 times.
- During these washes the instructors will return your elution fractions. Transfer 25 μL of each elution into a fresh 1.5 mL eppendorf tube for Part #3.
- Add your purified FKBP12 solution (both elutions, ~2 mL) to the desalting column. Change out the 15 mL conical for a fresh 15 mL tube then centrifuge for 2 min at 1000 g.
- Your desalted FKBP12 will be the flowthrough in the 15 mL conical.
Part 2: Concentrate FKBP12 protein solution
In order to use the purified and dialyzed FKBP12 protein in downstream assays, we will need to concentrate the amount of protein in solution. For this, we will use a centrifugal filter having a 3 kDa cutoff. The sample is first pipetted onto the filter, centrifuged to remove a fraction of fluid, and then recovered in a smaller volume using a pipet.
- Retrieve a 4 mL centrifugal filter unit from the front bench (it looks like a 15 mL conical tube with a filter unit inside of it).
- Pipette your desalted FKBP12 solution into the filter unit and screw on the cap.
- Make sure to mark the tube with your team color.
- Give the tube to the teaching faculty. All the filter units for the lab section will be spun together in the centrifuge at 4500 rpm for 30 minutes.
- During centrifugation, prepare a 1.5 mL eppendorf tube, clearly labeled with your team name (a colored sticker with TR or WF) as well as the contents (FKBP12).
- Once the centrifugation is complete, use a pipette to retrieve the remaining fluid on the filter and place in the labeled eppendorf tube.
- Use a P200 pipet tip to ensure all the fluid is retrieved from the filter unit.
- Transfer 25 μL of the concentrated protein solution to a fresh eppendorf tube for Part #3.
- Make sure to give your final tube of concentrated protein to the Teaching Faculty. It will be stored at 4 °C until your next experiment.
Part 3: Visualize purified protein with polyacrylamide gel electrophoresis (PAGE)
During the previous laboratory session, you reserved an aliquot of your induced and the uninduced cell lysate. In addition, the flow-through from the wash steps was stored. Today you will use SDS-PAGE to visualize the effectiveness of IPTG induction and the purification procedure.
- Retrieve the 15 μL aliquots of your induced and the uninduced cell lysates you prepared during the previous laboratory session. In addition, collect the flow-through from your wash steps and your purified, dialyzed protein solution.
- Transfer 25 μL from each of the wash flow-through samples into labeled 1.5 mL eppendorf tubes.
- Add 5 μL of Laemmli sample buffer to all of the samples prepared for SDS-PAGE
- This should include the uninduced / induced cell lysates, wash flowthroughs, elutions (from Part #2) and concentrated protein (from Part #2).
- Boil all samples for 5 min in the water bath located in the chemical fume hood.
- Secure the caps with the cap-locks located in the fume hood to ensure that the eppendorf caps do not pop open during the boiling step as this will result in your sample escaping the tube.
- If there is significant condensation at the lid of your tubes after boiling spin your samples before loading.
- You will load all samples and two molecular weight standards.
- A pre-stained ladder will be used to track the migration of your samples through the polyacrylamide gel.
- An unstained ladder with bands of known amounts of protein will be used to estimate protein concentration in your samples.
- Record the order in which you will load your samples and molecular weight standards in the polyacrylamide gel.
- When you are ready to load your samples, alert the teaching faculty.
- Please watch the demonstration closely to ensure your samples are correctly loaded and the polyacrylamide gel is not damaged during loading.
- Your samples will be electrophoresed at 200 V for 30-45 min.
- Following electrophoresis, use the spatula to carefully pry apart the plates that encase your polyacrylamide gel.
- Using wet gloves, transfer your polyacrylamide gel to a staining box and add enough dH2O to cover the gel.
- Wash the gel for 5 min at room temperature on the rotating table.
- Empty the water from the staining box in the sink.
- Be careful that the gel does not fall into the sink!
- Repeat Steps #12-13 a total of 3 times.
- Add 50 mL of BioSafe Coomassie to the staining box and incubate for 60 min at room temperature on the rotating table(or overnight depending on time).
- Empty the BioSafe Coomassie into the appropriate waste container in the chemical fume hood.
- Be careful that the gel does not fall into the waste container!
- Add 200 mL of dH2O to the staining box.
- Wash the gel for the remainder of the class on the rotating table.
- Replace the dH2O every 30 min.
Tomorrow the teaching faculty will transfer your gel to fresh dH2O and take a photograph. The image will be posted to the Class Data page.
Part 4: Measure protein concentration
SDS-PAGE enables you to visualize the presence of protein and provides information concerning the purity of your protein sample(s). Though comparing SDS-PAGE band intensities between samples and molecular weight standards gives an estimate of protein concentration, using a standard curve generated from samples of known protein concentration is a more precise method for measuring protein concentration.
Part 4a: Prepare diluted albumin (BSA) standards
- Obtain a 100 μL aliquot of 2.0 mg/mL albumin standard stock (labeled BSA) and a conical tube of PBS from the front bench.
- Prepare your standards according to the table below using PBS as the diluent:
| Vial |
Volume of diluent (μL) | Volume (μL) and source of albumin (vial) | Final albumin concentration (μg/mL) |
|---|---|---|---|
| A | 700 | 100 of stock | 250 |
| B | 400 | 400 of A | 125 |
| C | 450 | 300 of B | 50 |
| D | 400 | 400 of C | 25 |
| E | 400 | 100 of D | 5 |
| F | 400 | 0 | 0 = blank |
Part 4b: Prepare Working Reagent (WR) and measuring protein concentration
- Use the following formula to calculate the volume of WR required: (# of standards + # unknowns) * 0.215 = total volume of WR (in mL).
- Prepare the calculated volume of WR by mixing 50 parts of BCA Reagent A with 1 part BCA Reagent B (50:1 or 50-fold dilution of B).
- For example, if your calculated total volume of WR is 100 mL, then mix 98 mL of A, and 2 mL of B.
- Prepare your WR in a 1.5ml microcentrifuge tube.
- Pipet 25 μL of each standard prepared in Part 3a and 25 μL of your desalted and concentrated FKBP12 protein into separate, wells of a flat bottom 96-well plate.
- Add 200 μL of the WR to each 25 μL aliquot of the standard OR FKBP12 protein sample (do not mix the standard and FKBP12).
- Cover your plate with plastic wrap and incubate at 37°C incubator in the back of the lab for 30min.
- During the incubation step, check on your SDS-PAGE gel.
- Cool the plate to room temperature.
- Notify the teaching faculty you need to take your samples to the spectrophotometer plate reader.
- Obtain the absorbance at 562 nm for each well. Subtract the measurement of the Blank standard (F, after adding working reagent and being incubated) from that of all other individual standard and unknown samples.
- Generate your standard curve by plotting the Blank-corrected absorbance 562 for each albumin standard (A-F, B-F, ..., F-F) vs. its concentration in μg/mL.
- Use the standard curve to determine the protein concentration of purified FKBP12 in your sample.
Reagents
- Zeba spin desalting column (7000 MWCO)
- Amicon Ultra-4 Centrifugal Filter (3000 MWCO)
- SDS-PAGE Gels, buffer and ladder from Bio-Rad
- 4-20% polyacrylamide gels in Tris-HCl
- TGS buffer (25 mM Tris, 192 mM glycine, 0.1% (w/v) SDS, pH 8.3)
- Biorad Kaleidoscope and unstained marker sizes here
- Unstained marker masses in manual linked here
- BioSafe Coomassie G-250 Stain (Bio-Rad)
- Pierce BCA protein assay (ThermoFisher Scientific)
- 6x Reducing Laemmli Sample Buffer (Boston BioProducts)
Next day: Test protein activity using peptidyl-prolyl cis-trans isomerase assay