Difference between revisions of "20.109(S17):Module 1"
From Course Wiki
Noreen Lyell (Talk | contribs) (→References) |
Noreen Lyell (Talk | contribs) (→Lab links: day by day) |
||
| Line 23: | Line 23: | ||
M1D5: [[20.109(S17):Scan slides to identify FKBP12 binders (Day5) |Scan slides to identify FKBP12 binders]]<br> | M1D5: [[20.109(S17):Scan slides to identify FKBP12 binders (Day5) |Scan slides to identify FKBP12 binders]]<br> | ||
M1D6: [[20.109(S17):Complete data analysis (Day6) |Complete data analysis]]<br> | M1D6: [[20.109(S17):Complete data analysis (Day6) |Complete data analysis]]<br> | ||
| − | M1D7: [[20.109(S17):Identify chemical structures common among FKBP12 binders |Identify chemical structures common | + | M1D7: [[20.109(S17):Identify chemical structures common among FKBP12 binders |Identify chemical structures common in FKBP12 binders]]<br> |
==Data== | ==Data== | ||
Revision as of 00:09, 31 January 2017
Contents
Module 1
Lecturer: Angela Koehler
Instructors: Noreen Lyell, Leslie McClain and Maxine Jonas
TA: Rob Wilson
Lab manager: Hsinhwa Lee
Overview
Lab links: day by day
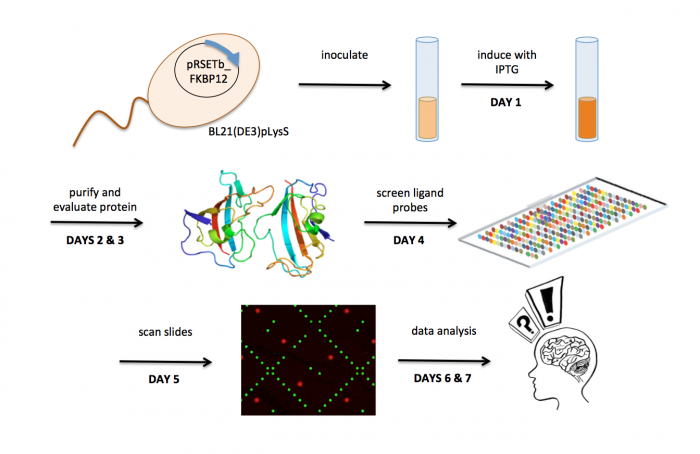
M1D1: In silico cloning and induce protein expression
M1D2: Purify induced protein
M1D3: Evaluate purity and concentration of protein
M1D4: Screen ligand library for FKBP12 binders
M1D5: Scan slides to identify FKBP12 binders
M1D6: Complete data analysis
M1D7: Identify chemical structures common in FKBP12 binders
Data
See all M1 student data on the Discussion page.
Assignments
Data summary
Mini-presentation
References
- A method for the covalent capture and screening of diverse small molecules in a microarray format. Nature Protocols. 1:2344-2352.
- Recent discoveries and applications involving small-molecule microarrays. Chemical Biology. 18:21-28.