20.109(F20):Module 2
Contents
Module 2
Lecturer: Jacquin Niles
Instructors: Noreen Lyell, Leslie McClain, and Becky Meyer
TAs:
Overview
REQUESTED WRITEUP FROM JACQUIN
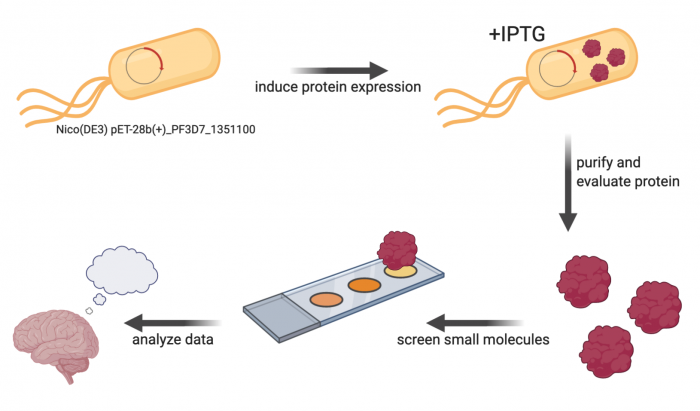
FROM KHAN: We have been looking for some small, essential proteins of unknown functions from P. falciparum for drug discovery attempts. We have cloned and knocked down multiple of the genes of interest in the parasite and in parallel, tried to express in E. coli. Some of them expressed in E. coli and if they are found to be essential by out parasite assays we go forward to do drug screening by SMM. The protein that we will be working on was one of them. A separate group has solved the crystal structure of this protein. The PDB ID is 1zso. However, despite having a structure in hand, we do not know much about this protein and literature is not rich for this gene. This is one of the troublesome feature of working with uncharacterized proteins but they can be good drug targets for at least because, most likely, these genes do not have a human orthologue. Therefore, drug selectivity can be achieved.
The primary experiment in this module is a small-molecule microarray (SMM), which ... To complete the SMM screen, the protein of interest must be cloned into an expression vector and then purified.
Research goal: ...
Lab links: day by day
M2D1: Complete in silico cloning of protein expression plasmid
M2D2: Perform protein purification protocol
M2D3: Assess purity and concentration of purified protein
M2D4: Prepare small molecule microarray (SMM) slides with purified protein
M2D5: Scan SMM slides to identify putative small molecule binders
M2D6: Analyze SMM data to confirm putative small molecule binders
M2D7: Examine putative small molecule binders for common features
Major assignments
Journal club presentation
Research article
References
A method for the covalent capture and screening of diverse small molecules in a microarray format. Nature Protocols. (2006) 1:2344-2352.
Recent discoveries and applications involving small-molecule microarrays. Chemical Biology. (2014) 18:21-28.