20.109(F20):Module 3
Contents
Module 3
Lecturer: Noreen Lyell
Instructors: Noreen Lyell, Leslie McClain, and Becky Meyer
TAs:
Overview
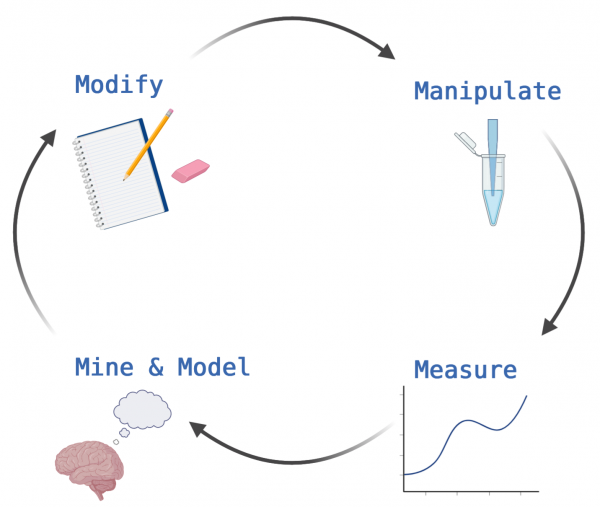
At the core of scientific discovery is building on the research of others to push the field forward. To accomplish this an iterative process is used to answer research questions: manipulate, measure, mine and model, and modify.
Manipulate: The first step in answering a research question is often designing and performing an experiment. The existing literature is an important resource at this stage as it provides information on what has already been done. In addition, the literature is useful in identifying which types experimental methods are appropriate for answering your specific research question. After the experiment is designed, the fun part of actually doing the benchwork can start.
Measure: It is not enough to just do an experiment. For the work to be meaningful, the data need to be collected and analyzed. The methods used for this stage in the process are just as important as those used for designing and performing the experiment. The type of analysis used is dependent on the experimental approach employed and on how the data are typically presented in the field. It is important that researchers in your same area of expertise are able to understand the data.
Mine and Model: After the data analysis is complete, it is time to figure out what the results mean in the context of the research question. As in the designing stage, here the existing literature is an important resource. The literature often provides context for your results. By exploring the literature it is possible to find links to your data that support the results.
Modify: Though the last stage in this list, this step really leads to a more refined research question...which will require you to design and perform a new experiment. Now that you have completed the stages of an experiment, here you use the results and information you gathered to improve the initial plan!
In Module 3 you will use this process to expand on the research completed by previous 109ers. In past iterations of this module students used a modified Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) system to modulate transcription in E. coli strain MG1655. Specifically, they used a CRISPR-based interference system (CRISPRi) to target genes within the anaerobic fermentative pathway in an effort to increase ethanol yield. Now it is your turn to continue the cycle of scientific discovery!
Research goal: Improve ethanol yield in E. coli MG1655 using the CRISPRi system by expanding upon previous research to design an optimized sgRNA targeting sequence.
Lab links: day by day
M3D1: Research project scope and experimental strategy
M3D2: Align gRNA sequences with genomic targets
M3D3: Analyze ethanol yield data
M3D4: Examine features in gRNA-targeted genomic sequences
M3D5: Design optimized gRNA sequence for enhanced ethanol production
Assignments
Research proposal presentation
Mini-report
References
CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nature Methods. (2013) 8: 2180-2196.
Metabolic engineering of Escherichia coli for production of mixed-acid fermentation end products. Frontiers in Bioengineering and Biotechnology. (2014) 2:1-12.