20.109(F20):Module 2
Contents
Module 2
Lecturer: Jacquin Niles
Instructors: Noreen Lyell, Leslie McClain, and Becky Meyer
TAs:
Overview
WRITEUP FROM JACQUIN REQUESTED
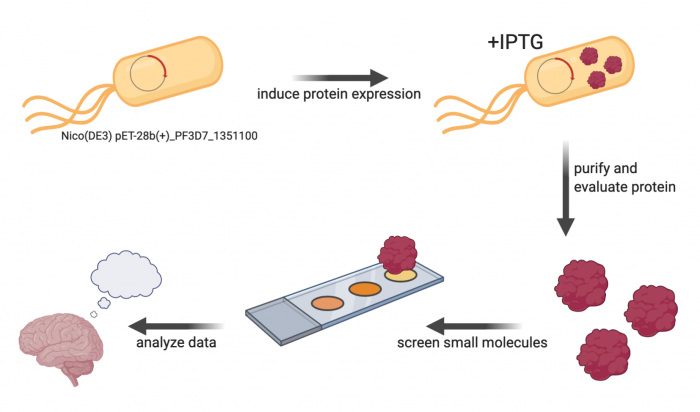
The primary experiment in this module is a small-molecule microarray (SMM), which ... To complete the SMM screen, the protein of interest must be cloned into an expression vector and then purified.
Research goal: Identify small molecules that putatively bind to the protein of interest.
Lab links: day by day
M2D1: Complete in silico cloning of protein expression plasmid
M2D2: Perform protein purification protocol
M2D3: Assess purity and concentration of purified protein
M2D4: Prepare small molecule microarray (SMM) slides with purified protein
M2D5: Scan SMM slides to identify putative small molecule binders
M2D6: Analyze SMM data to confirm putative small molecule binders
M2D7: Examine putative small molecule binders for common features
Major assignments
Journal club presentation
Research article
References
A method for the covalent capture and screening of diverse small molecules in a microarray format. Nature Protocols. (2006) 1:2344-2352.
Recent discoveries and applications involving small-molecule microarrays. Chemical Biology. (2014) 18:21-28.