Difference between revisions of "20.109(S24):M2D5"
Becky Meyer (Talk | contribs) |
Becky Meyer (Talk | contribs) (→Reagents list) |
||
| Line 103: | Line 103: | ||
==Reagents list== | ==Reagents list== | ||
| − | * | + | *Complete synthetic media - tryptophan (CSM-T) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.13 % amino acid mix lacking tryptophan (US Biological), 2% glucose (BD Bacto), 0.1% adenine hemisulfate (Sigma) |
| − | * | + | *Complete synthetic media - tryptophan + Cysteine, Methionine, and galactose (CSM-G) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.13 % amino acid mix lacking tryptophan (US Biologicals), 0.005% Methionine (Sigma), 0.005% Cysteine (Sigma), 2% galactose (Sigma), 0.1% adenine hemisulfate (Sigma) |
| − | + | *Cadmium nitrate (Sigma), stock concentration= 100mM | |
| − | *Cadmium | + | |
| − | + | ||
*ULTREX II, Ultrapure Nitric acid (J.T. Baker) | *ULTREX II, Ultrapure Nitric acid (J.T. Baker) | ||
*Hydrophilic PTFE 0.22μm filters (J.T. Baker) | *Hydrophilic PTFE 0.22μm filters (J.T. Baker) | ||
Revision as of 23:19, 12 March 2024
Contents
Introduction
Protocols
Part 1: Induce expression of YSD peptide
For timing reasons, the induction steps were completed prior to class. So you understand how the cell suspensions you will use for the metal uptake assay were created, please review the steps below.
- Inoculated 5 mL of SD-R media with a colony of ΔMet17 cells transformed with the pYAGA containing your peptide of interest.
- Incubated the culture overnight at 30 °C with shaking at 220 rpm.
- Dilute the overnight culture 1:10 in 10 mL of fresh SD-R media.
- Incubate at 30 °C for 4 hours with shaking at 220 rpm.
- To induce cell surface peptide expression, pellet cells and resuspend in 10ml of SD-G media.
- Incubate overnight at at 30 °C with shaking at 220 rpm.
Part 2: Stain transformed yeast to determine YSD expression
Prior to this session, the instructors transformed ...
Retrieve yeast culture labelled with your groups color from 30°C incubator
- Measure the optical density (OD) of your yeast on the spectrophotometer.
- You will have two samples: untransformed ΔMet17 yeast, and ΔMet17 yeast transformed with a pYAGA control plasmid expressing a peptide.
- Blank the spectrophotometer with >1000 uL of CSM in a clear cuvette
- Add 900 uL of CSM to a new cuvette. Add 100 uL of your yeast culture and mix well by pipetting up and down.
- Record the OD(@600nm) of your sample by taking a measurement using the spectrophotometer and multiplying by 10 (to account for the 10x dilution).
- Determine how many microliters of your sample are required to analyze 1x106 yeast cells.
- The conversion rate from OD to cells for yeast is: (107 yeast cells)/(OD x mL of culture)
- Thus, An optical density of 1 at 600nm equals 107 yeast cells per mL.
- Add the correct volume of culture (calculated above) such that 10^6 yeast are added to a microcentrifuge tube for each sample.
- Add an additional 900 uL of PBSA to each centrifuge tube and pellet the cells for 2 minutes in microcentrifuge at 4000xg.
- Discard supernatant carefully with vacuum aspirator or with pipette (Do not touch or disturb the yeast pellet!). Wash with an additional 1 mL of PBSA, pellet, and remove supernatant. Repeat washing step once more.
- Add antibody to samples: Add 1 uL of the antibody specific for the HA tag and conjugated to a 488 fluorophore to both tubes.
- Incubate samples on nutator at room temperature for 1 hour.
- During this incubation time, complete Part 3 of the protocol.
- After incubation, pellet cells at 4000xg for 2 minutes. Discard supernatant carefully with vacuum aspirator or with pipette (Do not touch or disturb the yeast pellet!). Wash with an additional 1 mL of PBSA, pellet, remove supernatant, and resuspend with 1 mL of PBSA.
- Place samples on ice until analysis.
In your laboratory notebook, complete the following:
- Calculate the volumes of yeast required to measure 1x106 cells
Part 3: Perform metal uptake experiment
- Obtain ΔMet17 transformed with pYAGA_peptide culture from the front bench
- While you prepare the experiment with your mutant, the instructors will prepare controls using the following cultures:
- Untransformed ΔMet17
- ΔMet17 transformed with control peptide
- Gently tritruate with a 1ml pipette to create a homogeneous suspension for each culture
- Obtain cuvettes from the front bench to measure the OD600 of each culture prior to beginning the experiment
- Add 1ml of each cell suspension to individual cuvettes and read on the spectrophotometer
- Use 1ml SD-G for a blank.
- Record these numbers in your notebook
- Using SD-G media as a diluent, dilute each of your cultures to OD600 ~ 1.0 and a final volume of 8ml. This does not have to be exact, but the cultures should have a similar OD600 before you begin the experiment.
- Add CdCl2 and FeCl2 for final concentration of 100μM in each culture and add to a new glass tube.
- Incubate your labeled glass tubes shaking at 30°C for 2.5 hours to allow uptake.
- During this incubation time, complete Parts 3 of the wiki.
- Following incubation, take an additional OD600 reading to account for any changes in culture density, and allow normalization of data across groups.
- Record this in your notebook.
- Transfer your cultures to 15ml conical tubes.
- Centrifuge cultures at 1000 xg for 5 minutes to pellet cells.
- During centrifugation, label 2 metal-free 15ml conical tubes for your mutant culture, and prepare a 10ml syringe filter for each culture
- Using serological pipette, remove 6ml media supernatent without disrupting the pellet and add it to a metal-free conical tube.
- Bring your samples to the front bench and add 175ul ultra-pure nitric acid to each sample. Return to your bench with your samples.
- Using a serological pipette, carefully triturate each sample to fully mix the acid.
- Open your 10ml filter syringe and place it over the top of a fresh metal-free conical tube. Add the mixed sample to the open syringe and use the plunger to push the sample through the 0.22 μM filter.
- Close each tube with sample and place them at the front bench.
- Discard materials used to filter the samples in the black bin at the front bench.
In your laboratory notebook, complete the following:
- What volume of CdCl2 will you add to obtain a final concentration of 100 μM?
- What volume of FeCl2 will you add to obtain a final concentration of 100 μM?
- Why is it important to use metal-free tubes to store your samples for analysis?
- Why is it important to filter your samples before submitting them for analysis?
Part 4: Accuri Flow Cytometry of Stained Yeast Samples
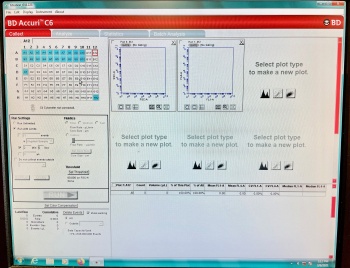
To prepare for analysis, the Accuri flow cytometer needs to be flushed with fluid to remove any contamination from previous samples. Your samples will then be filtered one by one into round bottom analyzer tubes and placed under the cytometer sample injection port (SIP). The machine will then suck up the sample, and produce the dot plots. The Accuri will record the data from each laser and filter for every cell. The gating that the instructor performs will simply help visualize the samples as we perform the experiment but all the data will be saved automatically for post-processing analysis.
- Turn machine on. MilliQ water (ultra-pure water) should be in a tube on the sample injection port (SIP).
- Visually check waste containers and buffers.
- Open BD AccuriC6 software on the computer.
- To clean the machine, place a round bottom tube with cleaning solution on the SIP and under Run settings, choose "Run with Limits" and set 5 minutes and click RUN.
- Once the 5min cleaning is complete, place an empty tube on the SIP and under Run settings, click the backflush button to rinse the system.
- Place a fresh tube with 2ml of MilliQ water and run for 2min.
- You are now ready to analyze your samples.
- Place one filter cap tube in a rack and take your unstained yeast sample from the ice bucket.
- Invert the yeast microcentrifuge tube once, take the P1000 pipet and take up the yeast sample. Place the pipet tip flush against the filter cap and push the yeast sample through the 35um mesh into the round bottom tube.
- Remove the filter cap off of the yeast sample, take the MilliQ water tube off of the SIP and place the tube with filtered yeast on the SIP.
- Choose the analysis parameters in the software under Run settings. Run with limits: 10,000 events, instead of a time limit.
- Choose an analysis cell in the software with no events recorded and hit the RUN button.
- Unused cells are white (Example, C1) and cells with stored data are blue (Example, A1).
- We will detail gating and fluorescent analysis on M2D5.
- As the analysis proceeds, prepare your next yeast sample according to the steps above.
- Data is collected in individual cells in the software. Make sure to move the event record to a new cell between samples. Hit RUN each time you start a new sample analysis.
- Either name the cells with experimental details or write in your notes which samples are analyzed in each cell.
- Once all of the samples are analyzed, place a round bottom tube of Decon solution (10% bleach) on the SIP and run for 2min.
- The flow cytometer is now ready for the next user.
In your laboratory notebook, complete the following:
- XXX
Reagents list
- Complete synthetic media - tryptophan (CSM-T) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.13 % amino acid mix lacking tryptophan (US Biological), 2% glucose (BD Bacto), 0.1% adenine hemisulfate (Sigma)
- Complete synthetic media - tryptophan + Cysteine, Methionine, and galactose (CSM-G) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.13 % amino acid mix lacking tryptophan (US Biologicals), 0.005% Methionine (Sigma), 0.005% Cysteine (Sigma), 2% galactose (Sigma), 0.1% adenine hemisulfate (Sigma)
- Cadmium nitrate (Sigma), stock concentration= 100mM
- ULTREX II, Ultrapure Nitric acid (J.T. Baker)
- Hydrophilic PTFE 0.22μm filters (J.T. Baker)
- Metal-Free sterile polypropylene tubes (VWR)
Next day: Quantify cadmium removal from media