Difference between revisions of "20.109(S20):Scan SMM slides to identify binders of TDP43 protein (Day5)"
Noreen Lyell (Talk | contribs) (→Introduction) |
Noreen Lyell (Talk | contribs) (→Protocols) |
||
| (21 intermediate revisions by one user not shown) | |||
| Line 4: | Line 4: | ||
==Introduction== | ==Introduction== | ||
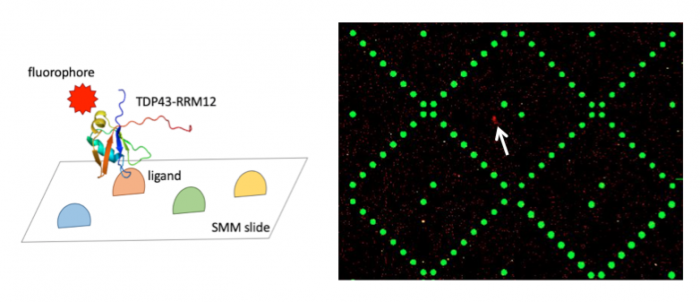
| − | In the previous laboratory session you | + | In the previous laboratory session you incubated SMM slides with your purified TDP43-RRM12 protein in an effort to identify ligand binders. To identify which of the small molecules, if any, you screened are able to putatively bind TDP43-RRM12, your slides were evaluated using a Genepix microarray scanner. The scanner measured the fluorescence signal emitted from each spot on the slide where a ligand was printed. Considering that neither the ligands nor TDP43-RRM12 protein is fluorescent, from where does this signal arise? |
| − | [[Image: | + | [[Image:Sp20 M1D5 SMM complex, scan.png|thumb|700px|center|]]<br> |
| − | + | The above image (right panel) is from a pilot experiment completed in preparation for this module. The green spots represent locations on the SMM slide where fluorescein was printed. As a reminder, fluorescein is a fluorescent dye used for alignment purposes. Correct alignment is critical to knowing which ligands are in which spots on the slide. Red spots are indicative of ligands that are bound by TDP43-RRM12 (example denoted by white arrow in right panel). The signal is due to the SNAP-Surface® Alexa Fluor® 647, a fluorophore that was conjugated to the TDP43-RRM12 protein via the SNAP sequence (schematic shown in left panel). | |
| − | When the | + | When the slides are imaged, the scanner exposes the SMM to excitation light specific to the fluorophores used in the experiment (''i.e.'' wavelengths that excite fluorescein and the Alexa Fluor 647 attached to TDP43-RRM12). The scanner then detects the intensity of the emitted fluorescence light to generate an image that can be analyzed. The intensity of the signal associated with putative ligand binders will be assessed in the next laboratory session. This analysis will provide a list of 'hits' that can be used in future studies. |
==Protocols== | ==Protocols== | ||
| − | ===Part 1 | + | ===Part 1: Scan slides=== |
| − | + | A researcher from Prof. Koehler's Laboratory will lead you through a demonstration on how to scan your SMM slides. You should take notes to ensure you know the purpose of each setting. An outline of the steps is included below for your reference. | |
| − | + | ||
| − | + | ||
| − | + | ||
#Place the slide in the scanner barcode side down. | #Place the slide in the scanner barcode side down. | ||
#Set the desired wavelengths that you want to scan. | #Set the desired wavelengths that you want to scan. | ||
#*For fluorescein: 532 nm | #*For fluorescein: 532 nm | ||
| − | #*For Alexa Fluor 647 | + | #*For Alexa Fluor 647: 635 nm |
#Set the wavelength, filter, PMT, and power settings. | #Set the wavelength, filter, PMT, and power settings. | ||
| − | #* | + | #*The Koehler Lab member will discuss these features during the demonstration. |
#*Record the purpose for each of these settings in your laboratory notebook! | #*Record the purpose for each of these settings in your laboratory notebook! | ||
#Run a preview scan to optimize the PMT for the 635 nm (red) emission. | #Run a preview scan to optimize the PMT for the 635 nm (red) emission. | ||
| Line 33: | Line 30: | ||
[[Image:Sp17 20.109 M1D5 Genepix scan.png|thumb|600px|center|]] | [[Image:Sp17 20.109 M1D5 Genepix scan.png|thumb|600px|center|]] | ||
| − | ===Part | + | ===Part 2: Review journal article=== |
| − | Skim and discuss the journal article by | + | Skim and discuss the journal article by Chen ''et al.'' titled "[[Media:SMMAlzABPeptide Koehler2010.pdf |Small molecule microarrays enable the discovery of compounds that bind the Alzheimer's αβ peptide]]" with your laboratory partner during the downtime that you will have today. |
| + | |||
| + | The initial experiment presented within this publication was an SMM that identified ligands binders of the αβ petptide. This first step is very similar to what you completed as part of Module 1. To further assess the results of the SMM, Chen ''et al.'' completed several follow-up experiments. In your laboratory notebook, list the experiments that were reported by the researchers and the conclusions of these experiments. You do not need to provide much detail here, just a sentence or two with what was done and what information/insight was gained. | ||
| + | |||
| + | When you complete your list, consider how these experiments are connected such that the paper provides a complete and coherent story. What is the story? | ||
| − | + | Use this list to guide a discussion with your partner concerning the organization of the data you will include in the Data summary. To get you started on your Data summary, create an outline that orders the figures that will be included in the assignment and record your thoughts on how the figures relate to each other (ie what is your story and how do the data support your story?). | |
| − | + | In addition, think about follow-up experiments that would be useful / insightful if you were to assess the putative binders identified in the SMM screen for TDP43-RRM12. How might you introduce / organize the follow-up experiments in the Implications and Future works section of your Data summary? For ideas on future works, refer to the M1D4 lecture notes. Also, consider the follow-up experiments completed by Chen ''et at.''. Keep in mind that not all of the approaches in the paper are relevant to your research! The questions that you address in your future works experiments should work toward a complete and coherent story. | |
==Reagents== | ==Reagents== | ||
| Line 45: | Line 46: | ||
==Navigation links== | ==Navigation links== | ||
| − | Next day: [[]] | + | Next day: [[20.109(S20):Analyze SMM data (Day6) |Analyze SMM data]]<br> |
| − | Previous day: [[]] | + | Previous day: [[20.109(S20):Perform small molecule microarray (SMM) with TDP43 protein (Day4) |Perform small molecule microarray (SMM) with TDP43 protein]] |
Latest revision as of 21:04, 21 February 2020
Contents
Introduction
In the previous laboratory session you incubated SMM slides with your purified TDP43-RRM12 protein in an effort to identify ligand binders. To identify which of the small molecules, if any, you screened are able to putatively bind TDP43-RRM12, your slides were evaluated using a Genepix microarray scanner. The scanner measured the fluorescence signal emitted from each spot on the slide where a ligand was printed. Considering that neither the ligands nor TDP43-RRM12 protein is fluorescent, from where does this signal arise?
The above image (right panel) is from a pilot experiment completed in preparation for this module. The green spots represent locations on the SMM slide where fluorescein was printed. As a reminder, fluorescein is a fluorescent dye used for alignment purposes. Correct alignment is critical to knowing which ligands are in which spots on the slide. Red spots are indicative of ligands that are bound by TDP43-RRM12 (example denoted by white arrow in right panel). The signal is due to the SNAP-Surface® Alexa Fluor® 647, a fluorophore that was conjugated to the TDP43-RRM12 protein via the SNAP sequence (schematic shown in left panel).
When the slides are imaged, the scanner exposes the SMM to excitation light specific to the fluorophores used in the experiment (i.e. wavelengths that excite fluorescein and the Alexa Fluor 647 attached to TDP43-RRM12). The scanner then detects the intensity of the emitted fluorescence light to generate an image that can be analyzed. The intensity of the signal associated with putative ligand binders will be assessed in the next laboratory session. This analysis will provide a list of 'hits' that can be used in future studies.
Protocols
Part 1: Scan slides
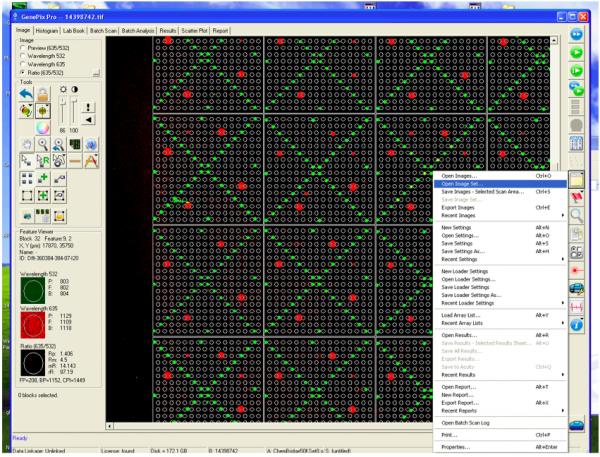
A researcher from Prof. Koehler's Laboratory will lead you through a demonstration on how to scan your SMM slides. You should take notes to ensure you know the purpose of each setting. An outline of the steps is included below for your reference.
- Place the slide in the scanner barcode side down.
- Set the desired wavelengths that you want to scan.
- For fluorescein: 532 nm
- For Alexa Fluor 647: 635 nm
- Set the wavelength, filter, PMT, and power settings.
- The Koehler Lab member will discuss these features during the demonstration.
- Record the purpose for each of these settings in your laboratory notebook!
- Run a preview scan to optimize the PMT for the 635 nm (red) emission.
- Complete a full scan using the optimal PMT value.
- Following the scan, you will see an image of your slide as below.
Part 2: Review journal article
Skim and discuss the journal article by Chen et al. titled "Small molecule microarrays enable the discovery of compounds that bind the Alzheimer's αβ peptide" with your laboratory partner during the downtime that you will have today.
The initial experiment presented within this publication was an SMM that identified ligands binders of the αβ petptide. This first step is very similar to what you completed as part of Module 1. To further assess the results of the SMM, Chen et al. completed several follow-up experiments. In your laboratory notebook, list the experiments that were reported by the researchers and the conclusions of these experiments. You do not need to provide much detail here, just a sentence or two with what was done and what information/insight was gained.
When you complete your list, consider how these experiments are connected such that the paper provides a complete and coherent story. What is the story?
Use this list to guide a discussion with your partner concerning the organization of the data you will include in the Data summary. To get you started on your Data summary, create an outline that orders the figures that will be included in the assignment and record your thoughts on how the figures relate to each other (ie what is your story and how do the data support your story?).
In addition, think about follow-up experiments that would be useful / insightful if you were to assess the putative binders identified in the SMM screen for TDP43-RRM12. How might you introduce / organize the follow-up experiments in the Implications and Future works section of your Data summary? For ideas on future works, refer to the M1D4 lecture notes. Also, consider the follow-up experiments completed by Chen et at.. Keep in mind that not all of the approaches in the paper are relevant to your research! The questions that you address in your future works experiments should work toward a complete and coherent story.
Reagents
- Genepix 4300 microarray scanner (Molecular Devices)
- Genepix Pro software (Molecular Devices)
Next day: Analyze SMM data