20.109(S20):Module 2
Contents
Module 2
Lecturers: Ernest Fraenkel and Noreen Lyell
Instructors: Noreen Lyell and Leslie McClain, and Becky Meyer
Research assistant: Anne Shen
TAs: Kevin Chung and Joe Kreitz
Lab manager: Hsinhwa Lee
Overview
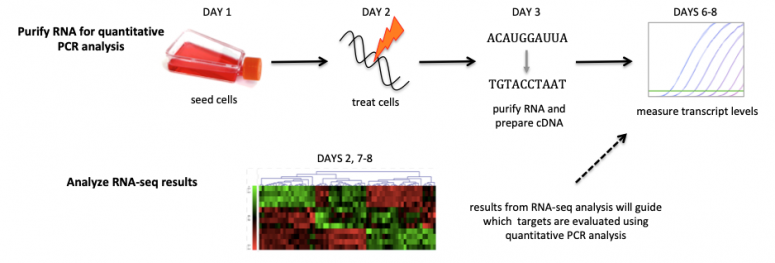
In Module 2 you will use two RNA-seq data sets to assess the effect of drug treatment on mammalian cells. RNA-seq is used to measure the transcriptome of a population of cells. By identifying which genes are active in response to a treatment, insights can be gathered regarding the cellular response. Specifically, the drug treatment you will use in your experiment is etoposide. Etoposide is known to generate double strand breaks in DNA and is used in chemotherapy. You will apply etoposide to DLD-1 cells, which are a cell line that originated from the colon of a male patient with colorectal adenocarcinoma Dukes Type C. In addition, you will compare the transcriptome results of your experiment (DLD-1 cells treated with etoposide) to published data using a different cell line (A549 cells) that were also treated with etoposide.
An additional goal of Module 2 is for you to synthesize data from different experiments into a single story. More often that not, researchers reach conclusions using results gathered from more than one experimental approach. Multiple layers of support can bolster a conclusion as they eliminate biases that may exist in a particular method of analysis. Furthermore, for complex questions it may be necessary to include data from several approaches to provide a sufficient conclusion.
Lab links: day by day
M2D1: Prepare cells for RNA purification and practice RNA-seq data analysis methods
M2D2: Treat cells with etoposide and analyze RNA-seq data
M2D3: Purify RNA from etoposide-treated cells and generate cDNA
M2D4: Journal club I presentations
M2D5: Journal club II presentations
M2D6: Analyze RNA-seq data and select gene targets for quantitative PCR experiment
M2D7: Review qPCR experiment and complete statistical analysis
M2D8: Complete RNA-seq data analysis
Assignments
Journal club presentation
Research article