20.109(S17):Examine transcript levels in response to DNA damage (Day 4)
From Course Wiki
Revision as of 02:18, 3 February 2017 by Noreen Lyell (Talk | contribs)
Contents
Introduction
Real-time reverse transcription-polymerase chain reaction (RRT-PCR) allows researchers to monitor the results of PCR as amplification is occurring (this technique is also referred to as quantitative or real-time PCR). During RRT-PCR data are collected throughout the amplification process using a fluorescent dye. The fluorescent dye is highly specific for double-stranded DNA and when bound to DNA molecules the fluorescence intensity increases proportionate to the increase in double-stranded product. In contrast, the data for traditional PCR are simply observed as a band on a gel (remember back to M1D5).
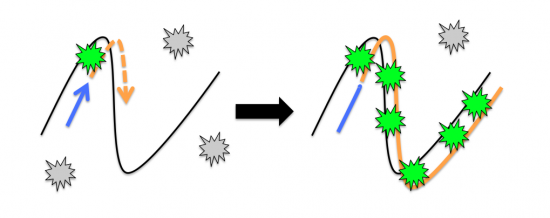
The fluorescent dye binds to double-stranded DNA during the cycles of PCR. At the annealing temperature the primer (blue arrow) binds to the template (black line). During an incubation at the extension temperature the new copy of DNA (orange dashed arrow) is sythesized by the polymerase enzyme. The inactive fluorescent dye molecules present in the reaction (grey stars) bind to the newly generated double-stranded DNA and become activated (green stars). To eliminate clutter, the basepairs between the DNA strands were omitted. An animation of this process is linked here.
Protocols
Part 1: Setup quantitative PCR assay
Part 2: Discussion of Research article details
Reagents
Next day: Journal club I
Previous day: Assess cell survival and harvest RNA for quantitative PCR assay