Difference between revisions of "20.109(S17):Confirm cell lines and practice tissue culture techniques (Day1)"
Noreen Lyell (Talk | contribs) (→Part 1b: Seed cells for survival assay) |
Noreen Lyell (Talk | contribs) (→Part 2c: Prepare cell lysates for polyacrylamide gel electrophoresis (PAGE)) |
||
| (41 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
==Introduction== | ==Introduction== | ||
| − | To be confident | + | To be confident in your findings related to DNA repair, you will first validate the cell lines you are using to assess the effect of drug treatment following DNA damage on NHEJ. It is important to confirm the cells you are using are indeed mutants in the BRCA2 protein to interpret the data you collect in this module. You will do this by completing a Western blot. A Western blot allows researchers to detect specific proteins within a mixed sample, such as cell lysate. |
| − | [[Image:Sp17 20.109 M2D1 etoposide effect.png|thumb|300px|right|Image modified from Hucl et al. [http://www.ncbi.nlm.nih.gov/pubmed/18593900 PMID:18593900] ]]We will use this protein analysis technique to assess BRCA2 levels in the wild-type DLD-1 and mutant DLD-1 BRCA2(-/-) (referred to as BRCA2-) cell lines. The wild-type or 'parental' DLD-1 cell line originated from | + | [[Image:Sp17 20.109 M2D1 etoposide effect.png|thumb|300px|right|Image modified from Hucl et al. [http://www.ncbi.nlm.nih.gov/pubmed/18593900 PMID:18593900] ]]We will use this protein analysis technique to assess BRCA2 levels in the wild-type DLD-1 and mutant DLD-1 BRCA2(-/-) (referred to as BRCA2-) cell lines. The wild-type or 'parental' DLD-1 cell line originated from the colon of a male patient with colorectal adenocarcinoma Dukes Type C. The BRCA2- mutant cell line was created via disruption of BRCA2 exon 11. This mutation results in cells that are deficient in DNA repair. In the image to the right, cell survival decreased as higher concentrations of a DNA damaging agent, etoposide, were added. |
You will work with these cells, mostly, in our tissue culture room. Tissue culture was developed ~100 years ago as a means to study mammalian biology, and since that time we have learned a tremendous amount by observing the behavior of mammalian cells maintained in the laboratory. The term tissue culture was originally coined because researchers were doing exactly that, extracting tissue and letting it live in a dish for a short time. Today, most tissue culture experiments are done using cells rather than tissues. Much of what we know about cancer, heritable diseases, and the effects of the environment on human health has been derived from studies of cultured cells. | You will work with these cells, mostly, in our tissue culture room. Tissue culture was developed ~100 years ago as a means to study mammalian biology, and since that time we have learned a tremendous amount by observing the behavior of mammalian cells maintained in the laboratory. The term tissue culture was originally coined because researchers were doing exactly that, extracting tissue and letting it live in a dish for a short time. Today, most tissue culture experiments are done using cells rather than tissues. Much of what we know about cancer, heritable diseases, and the effects of the environment on human health has been derived from studies of cultured cells. | ||
| Line 11: | Line 11: | ||
What types of cells do people study, and from where do they come? Cells acquired directly from animal tissue are called primary cells. They are difficult to culture, largely because primary cells in this context divide only a limited number of times. This limitation on the lifespan of cultured primary cells, called the Hayflick limit, is a problem because it requires a researcher to repeatedly remove tissues from animals to complete a study. Cell isolation processes can be quite labor-intensive, and also can complicate data analysis due to inherent animal-to-animal variation. To get around this problem, researchers study cells that are immortal, which means the cells are able to divide indefinitely. Though using immortal cells is preferable for many reasons, some inherent cell-to-cell variation still exists in such populations and the genetic changes that cause immortality may affect experimental outcomes. | What types of cells do people study, and from where do they come? Cells acquired directly from animal tissue are called primary cells. They are difficult to culture, largely because primary cells in this context divide only a limited number of times. This limitation on the lifespan of cultured primary cells, called the Hayflick limit, is a problem because it requires a researcher to repeatedly remove tissues from animals to complete a study. Cell isolation processes can be quite labor-intensive, and also can complicate data analysis due to inherent animal-to-animal variation. To get around this problem, researchers study cells that are immortal, which means the cells are able to divide indefinitely. Though using immortal cells is preferable for many reasons, some inherent cell-to-cell variation still exists in such populations and the genetic changes that cause immortality may affect experimental outcomes. | ||
| − | The art of tissue culture lies in the ability to create conditions that are similar to what a cell would experience in an animal, namely 37°C and neutral pH. Blood nourishes the cells in an animal, and blood components are | + | The art of tissue culture lies in the ability to create conditions that are similar to what a cell would experience in an animal, namely 37°C and neutral pH. Blood nourishes the cells in an animal, and blood components are therefore used to feed cells in culture. Serum, the cell-free (and clotting-factor free) component of blood, contains many of the factors necessary to support the growth of cells outside the animal. Consequently, serum is frequently added to tissue culture medium, although serum-free media (also called chemically defined media) exist and support some types of cultured cells. |
| − | Cultured mammalian cells must grow in a germ-free environment and researchers using tissue culture must be skilled in sterile or | + | Cultured mammalian cells must grow in a germ-free environment and researchers using tissue culture must be skilled in sterile or aseptic technique. Bacteria double very quickly relative to mammalian cells. An average mammalian cell doubles about once per day whereas many common bacteria can double every 20 minutes under optimal conditions. Consequently, if you were to co-culture 100 mammalian cells and 1 bacteria together in a flask, within 24 hours you would have ~200 unhappy mammalian cells, and about 100 million happy bacteria! Needless to say, you would not find it very useful to continue to study the behavior of your mammalian cells under these conditions. |
| − | Today you will learn and practice | + | Today you will learn and practice aseptic technique to collect and count cells that were previously cultured by the teaching faculty. You will use the cells you collect to seed (or culture) cells for a survival assay that will assess the effect of drug treatment following DNA damage. To ensure you are well-prepared for your work in the tissue culture room, your instructor will provide a demonstration before you begin. Because the tissue culture room is not large enough to accommodate the entire class at once, we will be split into two groups today... and all subsequent days that involve tissue culture. Half of the class will begin in tissue culture with Part 1 and the other half of the class will begin by preparing previously seeded DLD-1 and BRCA2- cells for Western blot analysis. Your instructor will designate the groups. |
==Protocols== | ==Protocols== | ||
| Line 33: | Line 33: | ||
#Clean tissue culture hood. | #Clean tissue culture hood. | ||
| − | ====Part 1b: Seed cells for | + | ====Part 1b: Seed cells for RNA purification==== |
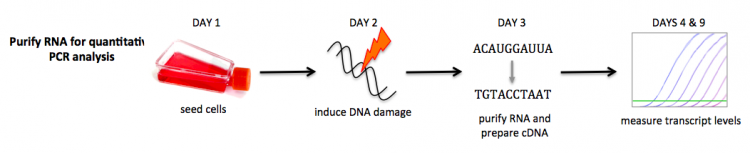
| − | You will use the techniques you learned during the demonstration to seed | + | You will use the techniques you learned during the demonstration to seed T75 flasks with cells. These cells will be used to assess the effect of DNA damage on gene expression. More specifically, you will examine this effect in the BRCA2- mutant cells compared to the DLD-1 wild-type cells. |
| − | [[Image:Sp17 20.109 | + | [[Image:Sp17 20.109 RNA purification R2.png|thumb|750px|center|]] |
| − | Each team will receive two | + | Each team will receive two T75 culture flasks: one with DLD-1 (wild-type) cells, and one with BRCA2- mutant cells. You will enzymatically detach these adherent cells, count them, and seed them at specific densities in T75 flasks. |
<font color=red>'''It is essential that you do not mix up or cross-contaminate the DLD-1 and BRCA2- cell lines! We suggest that one partner is responsible for each flask, and that partners do not share Pasteur pipets or other equipment.'''</font color=red> | <font color=red>'''It is essential that you do not mix up or cross-contaminate the DLD-1 and BRCA2- cell lines! We suggest that one partner is responsible for each flask, and that partners do not share Pasteur pipets or other equipment.'''</font color=red> | ||
| Line 53: | Line 53: | ||
#After you look at your cells, take the flask to your tissue culture hood to begin the seeding procedure. | #After you look at your cells, take the flask to your tissue culture hood to begin the seeding procedure. | ||
#Aspirate the media from the cells using a sterile Pasteur pipet. | #Aspirate the media from the cells using a sterile Pasteur pipet. | ||
| − | #Wash the cells by adding | + | #Wash the cells by adding 5 mL PBS using a 5 mL pipet. Slightly tip the flask back and forth to rinse the cells then aspirate the PBS with a '''fresh''' Pasteur pipet. |
#To dislodge the cells from the flask, you will add trypsin, a proteolytic enzyme. | #To dislodge the cells from the flask, you will add trypsin, a proteolytic enzyme. | ||
| − | #*With a 2 mL pipet, add | + | #*With a 2 mL pipet, add 2 mL of trypsin to the flask. |
#*2 mL pipets are tricky! They fill up quickly. '''Be careful not to pull up the liquid too quickly or it will go all the way up your pipet into the pipet-aid!''' If this happens, please alert the teaching faculty rather than returning the pipet-aid to the rack. | #*2 mL pipets are tricky! They fill up quickly. '''Be careful not to pull up the liquid too quickly or it will go all the way up your pipet into the pipet-aid!''' If this happens, please alert the teaching faculty rather than returning the pipet-aid to the rack. | ||
#Tip the flask in each direction to distribute the trypsin evenly then incubate the cells at 37°C for 10 minutes using a timer. | #Tip the flask in each direction to distribute the trypsin evenly then incubate the cells at 37°C for 10 minutes using a timer. | ||
#*This is a great time to clear out your trash and read ahead! | #*This is a great time to clear out your trash and read ahead! | ||
| − | #Retrieve your flasks from the incubator and firmly tap the bottom | + | #Retrieve your flasks from the incubator and firmly tap the bottom 10 times to dislodge the cells. |
#*Check your cells using the microscope to ensure they are dislodged. They should appear round and move freely. | #*Check your cells using the microscope to ensure they are dislodged. They should appear round and move freely. | ||
| − | #*If your cells are not detached from the flask, incubate at 37°C for an additional minute. | + | #*If your cells are not detached from the flask, incubate at 37 °C for an additional minute. |
| − | #When your cells are dislodged, move your flask back into the tissue culture hood and add | + | #When your cells are dislodged, move your flask back into the tissue culture hood and add 5 mL of media to the cells then pipet the liquid up and down (“triturate”) to break up cells that are clumped together and suspend them in the liquid. |
#*'''Note: do not take up or release all the liquid, in order to avoid bubbles.''' | #*'''Note: do not take up or release all the liquid, in order to avoid bubbles.''' | ||
#Transfer the suspended cells into a labeled 15 mL conical tube | #Transfer the suspended cells into a labeled 15 mL conical tube | ||
| Line 77: | Line 77: | ||
'''Seeding cells''' | '''Seeding cells''' | ||
| − | # | + | #Obtain four T75 flasks and label two DLD-1 and two BRCA2-. |
#*Include your section information and the date. | #*Include your section information and the date. | ||
| − | #Calculate the volume of each cell suspension you need to seed | + | #Calculate the volume of each cell suspension you need to seed 100,000 DLD-1 cells and 250,000 BRCA2- cells per T75 flask. |
| − | #*For example, if your concentration is 1 million (1 M) cells/mL, you would | + | #*For example, if your concentration is 1 million (1 M) cells/mL, you would seed the fresh T75 flasks 250 μL of DLD-1 cells. |
| − | #Add | + | #Subtract the volume of cell suspension that you calculated in Step #2 from 12 mL. |
| + | #*Add this volume of media to the fresh T75 flask. | ||
#Add the volume of cell suspension that you calculated in Step #2. | #Add the volume of cell suspension that you calculated in Step #2. | ||
| − | #*You should mix your cell suspension by pipetting before distributing the small volume of cells to the | + | #*You should mix your cell suspension by pipetting before distributing the small volume of cells to the flasks as the cells likely settled to the bottom of the conical while you were counting. |
| − | #Finally, tilt your | + | #Finally, tilt your flasks back and forth to distribute the cells evenly. |
| + | #Before moving your flasks to the 37 °C incubator, use the microscope to visually confirm that cells are present. | ||
| − | '''Cleaning the tissue culture hood''' | + | '''Cleaning the tissue culture hood''' <br> |
| − | The next group who uses your hood should find the surfaces wiped down and free of equipment. | + | The next group who uses your hood should find the surfaces wiped down and free of equipment. |
#Aspirate any remaining cell suspensions. | #Aspirate any remaining cell suspensions. | ||
| − | #Dispose of all vessels that held cells in the biohazard waste box and be sure that all sharps are in the | + | #Dispose of all vessels that held cells in the biohazard waste box and be sure that all sharps are in the sharps jar. |
| − | #Remove any equipment that you transferred into the hood and return | + | #Remove any equipment or supplies that you transferred into the hood and return to the appropriate location. |
| + | #*'''Please leave the equipment that was already there.''' | ||
#Spray the TC hood surface with 70% ethanol and wipe with paper towels. | #Spray the TC hood surface with 70% ethanol and wipe with paper towels. | ||
#*Be sure the paper towels are disposed of in the biohazard waste box! | #*Be sure the paper towels are disposed of in the biohazard waste box! | ||
| Line 96: | Line 99: | ||
===Part 2: Begin Western blot=== | ===Part 2: Begin Western blot=== | ||
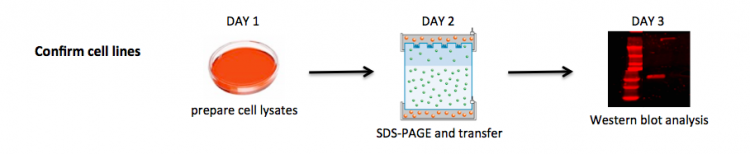
| − | [[Image:Sp17 20.109 | + | You will use cells seeded by the teaching faculty to prepare cell lysates that will be used to confirm that BRCA2 is indeed absent from the mutant cell line, and present in the parental cell line. |
| + | |||
| + | [[Image:Sp17 20.109 confirm cells R2.png|thumb|750px|center|]] | ||
====Part 2a: Prepare cell lysates==== | ====Part 2a: Prepare cell lysates==== | ||
| Line 108: | Line 113: | ||
#Aspirate the media from each well by holding the tip of the pasteur pipet (be sure to cap the pasteur pipet with a clean yellow tip!) at the bottom of the well. | #Aspirate the media from each well by holding the tip of the pasteur pipet (be sure to cap the pasteur pipet with a clean yellow tip!) at the bottom of the well. | ||
#Add 2 mL of ice-cold PBS to each dish. | #Add 2 mL of ice-cold PBS to each dish. | ||
| − | |||
#Aspirate the ice-cold PBS – '''be sure to remove ALL of the PBS after this wash.''' | #Aspirate the ice-cold PBS – '''be sure to remove ALL of the PBS after this wash.''' | ||
#Add 100 μL of RIPA lysis buffer to the top of each dish and allow it to run down the dish to the bottom. | #Add 100 μL of RIPA lysis buffer to the top of each dish and allow it to run down the dish to the bottom. | ||
| Line 117: | Line 121: | ||
#*At this step it is important to make sure there are no aggregates or clumps of cell lysate remaining in the dish. Do this by tilting the plate so that light reflects off the bottom. If you see any 'chunks', pipette your lysate back into the dish to dissolve the chunk, then re-transfer everything into the eppendorf tube. | #*At this step it is important to make sure there are no aggregates or clumps of cell lysate remaining in the dish. Do this by tilting the plate so that light reflects off the bottom. If you see any 'chunks', pipette your lysate back into the dish to dissolve the chunk, then re-transfer everything into the eppendorf tube. | ||
#Incubate the eppendorf tubes with your cell lysates on ice for 10 min. | #Incubate the eppendorf tubes with your cell lysates on ice for 10 min. | ||
| − | # | + | #Toward the end of this incubation bring your tubes, on ice, to the front bench to sonicate the lysate for 10seconds, followed by 30 seconds on ice. Repeat this 3 times. |
#Spin the cell lysate at maximum speed in the cold room centrifuge for 10 min to pellet insoluble material. | #Spin the cell lysate at maximum speed in the cold room centrifuge for 10 min to pellet insoluble material. | ||
| − | #*The teaching faculty will | + | #*The teaching faculty will take you to the centrifuge when you are ready. |
| + | #*Meanwhile, label two fresh eppendorf tubes and add them to your ice bucket to chill for a later step. | ||
#*This step is typically referred to as "clearing" the lysate. The pellet at the bottom contains the DNA from the cell, and genomic DNA can get very soupy making it difficult to load your lysate on the SDS-PAGE gel. | #*This step is typically referred to as "clearing" the lysate. The pellet at the bottom contains the DNA from the cell, and genomic DNA can get very soupy making it difficult to load your lysate on the SDS-PAGE gel. | ||
#Transfer the supernatant to the fresh eppendorf tubes – be careful not to disturb the pellet at the bottom! | #Transfer the supernatant to the fresh eppendorf tubes – be careful not to disturb the pellet at the bottom! | ||
| Line 127: | Line 132: | ||
You will now measure the total protein concentration in each cell lysate to determine the volume that you will use for the polyacrylamide gel separation. | You will now measure the total protein concentration in each cell lysate to determine the volume that you will use for the polyacrylamide gel separation. | ||
| − | # | + | #Prepare your assay reagent by mixing 1 part BioRad dye with 4 parts deionized water. |
| − | #*Prepare a 'blank' by adding | + | #*You will need 1 mL of diluted dye per sample, this will be enough for your blank and two cell lysate samples. |
| − | # | + | #*This reagent is hazardous chemical waste. Teaching staff will collect all used and unused samples once measurements are complete. |
| − | # | + | # Prepare a 'blank' by adding 1 mL of diluted dye then 1 μL of your leftover RIPA buffer to a cuvette. Cover the cuvette with parafild then invert to mix. |
| − | #* | + | #Add 1 mL of diluted dye then 1 μL of each cell lysate to a plastic cuvette. As above, cover with parafilm and invert to mix. |
| − | # | + | #Incubate cuvettes with dye plus sample for a minimum of 5 min at room temperature. |
| − | #*Use the RIPA sample to blank the spectrophotometer. | + | #*Samples should incubate at room temperature for no more than 1 h. |
| − | #Calculate the | + | #Measure the absorbance of each sample at 595 nm. |
| − | #* | + | #*Use the RIPA sample to blank the spectrophotometer prior to measuring samples. |
| − | + | #Calculate the protein concentrations of your DLD-1 and BRCA2- cell lysate using the following information: | |
| − | + | #*Standard curve linear regression: y = 0.058*x - 0.018, where y is your measured absorbance and x is your sample protein concentration in μg/μL. | |
| − | #Next, calculate the volumes of lysate and water required to add | + | #Next, calculate the volumes of lysate and water required to add 40 μg of total protein to the polyacrylamide gel in a total volume of 20 μL, per lysate sample. |
| − | #*If your concentration is greater than | + | #*If your concentration is greater than 2 μg/μL, use water to make up the remaining volume. |
| − | #*If your concentration is less than | + | #*If your concentration is less than 2 μg/μL for at least one sample, scale both samples down to a lower protein amount, such as 20 μg. |
#'''Do not throw away the remainder of your cell lysate!''' We will store these samples at -80 °C in case your Western blot needs to be repeated. | #'''Do not throw away the remainder of your cell lysate!''' We will store these samples at -80 °C in case your Western blot needs to be repeated. | ||
| − | ====Part 2c: | + | ====Part 2c: Prepare cell lysates for polyacrylamide gel electrophoresis (PAGE)==== |
| − | + | #Add 4 μL of 6X Laemmli sample buffer to the cell lysate samples you prepared in Part 2b Step #7. | |
| − | + | ||
| − | #Add 4 μL of 6X Laemmli sample buffer to | + | |
#Briefly vortex each tube, then centrifuge to collect the samples at the bottom of the tubes. | #Briefly vortex each tube, then centrifuge to collect the samples at the bottom of the tubes. | ||
#Put lid locks on the eppendorf tubes and boil for 5 minutes in the water bath that is in the fume hood. | #Put lid locks on the eppendorf tubes and boil for 5 minutes in the water bath that is in the fume hood. | ||
| − | # | + | #Allow your samples to cool to room temperature, then give them to the teaching faculty for storage at -20 °C until your next laboratory session. |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ===Part 3: Review research paper=== | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | We will end today with a discussion of the Dietlein ''et al.'' [[Media:M2D7 Cancer Discovery-2014-Dietlein-592-605.pdf |research article]]. In their research, the authors completed a screen to examine 1319 cancer-associated genes from 67 cell lines to identify cancer-cell specific mutations that are associated with DNA-PKcs dependence or addiction. Refer to the [[20.109(S17):Module_2#Overview| M2 overview]] for more information on addiction in cancer cells. | |
| − | + | ||
| − | + | Our paper discussion will assist you in writing a cohesive story that clearly reports the data and provides strong support for the conclusions made about the data. '''During the paper discussion, everyone is expected to participate - either by volunteering or by being called upon!''' | |
| − | + | ||
| − | + | '''Introduction''' | |
| − | + | ||
| − | + | Remember the key components of an introduction: | |
| − | + | *What is the big picture? | |
| − | + | *Is the importance of this research clear? | |
| − | + | *Are you provided with the information you need to understand the research? | |
| − | + | *Do the authors include a preview of the key results? | |
| − | + | ||
| − | + | '''Results''' | |
| − | + | ||
| − | + | Carefully examine the figures. First, read the captions and use the information to 'interpret' the data presented within the image. Second, read the text within the results section that describes the figure. | |
| − | + | *Do you agree with the conclusion(s) reached by the authors? | |
| − | + | *What controls are included and are they appropriate for the experiment performed? | |
| − | + | *Are you convinced that the data are accurate and/or representative? | |
| − | + | ||
| − | + | '''Discussion''' | |
| − | + | ||
| − | + | Consider the following components of a discussion: | |
| − | + | *Are the results summarized? | |
| + | *Did the authors 'tie' the data together into a cohesive and well-interpreted story? | ||
| + | *Do the authors overreach when interpreting the data? | ||
| + | *Are the data linked back to the big picture from the introduction? | ||
==Reagents== | ==Reagents== | ||
| + | |||
| + | From Thermo Fischer | ||
| + | *DLD-1 wild-type cells | ||
| + | *BRCA2-/- mutant cells | ||
| + | |||
| + | Media components from Life Technologies, Inc. unless noted otherwise. | ||
| + | |||
| + | *DLD-1 and BRCA2-/- cell media | ||
| + | **RPMI 1640 | ||
| + | **10% fetal bovine serum (FBS) | ||
| + | **100X antibiotic solution from Cellgro | ||
| + | ***10,000 U/mL Penicillin | ||
| + | ***10,000 U/mL Streptomycin | ||
| + | *phosphate-buffered saline (PBS) | ||
| + | *0.05% trypsin/0.91 mM EDTA | ||
| + | *Incubator maintains 37°C, 5% CO2 and 95% relative humidity | ||
| + | |||
| + | From Boston Bioproducts: | ||
| + | *RIPA lysis buffer | ||
| + | ** 50 mM Tris-HCl, pH 7.4 | ||
| + | ** 150 mM NaCl | ||
| + | ** 1% NP-40 | ||
| + | ** 0.5% sodium deoxycholate | ||
| + | ** 0.1% SDS | ||
| + | *100X protease inhibitor cocktail | ||
| + | ** AEBSF | ||
| + | ** Aprotinin | ||
| + | ** E-64 Besstain Leupeptin | ||
| + | ** EDTA | ||
| + | *6x reducing Laemmli sample buffer | ||
| + | ** 375 mM Tris HCl, pH 6.8 | ||
| + | ** 9% SDS | ||
| + | ** 50% glycerol | ||
| + | ** 9% betamercaptoethanol | ||
| + | ** 0.03% bromophenol blue | ||
| + | |||
| + | From Bio-Rad | ||
| + | *Protein Assay Dye (5X) | ||
==Navigation links== | ==Navigation links== | ||
| − | Next day: [[20.109(S17): | + | Next day: [[20.109(S17):Prepare Western blot and induce DNA damage for quantitative PCR assay (Day2)| Prepare Western blot and induce DNA damage for quantitative PCR assay]] |
Previous day: [[20.109(S17):Identify chemical structures common among FKBP12 binders |Last day of M1]] | Previous day: [[20.109(S17):Identify chemical structures common among FKBP12 binders |Last day of M1]] | ||
Latest revision as of 21:06, 9 March 2017
Contents
Introduction
To be confident in your findings related to DNA repair, you will first validate the cell lines you are using to assess the effect of drug treatment following DNA damage on NHEJ. It is important to confirm the cells you are using are indeed mutants in the BRCA2 protein to interpret the data you collect in this module. You will do this by completing a Western blot. A Western blot allows researchers to detect specific proteins within a mixed sample, such as cell lysate.
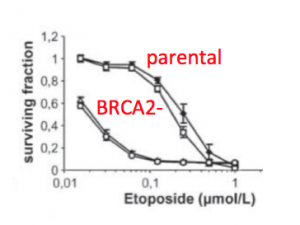
You will work with these cells, mostly, in our tissue culture room. Tissue culture was developed ~100 years ago as a means to study mammalian biology, and since that time we have learned a tremendous amount by observing the behavior of mammalian cells maintained in the laboratory. The term tissue culture was originally coined because researchers were doing exactly that, extracting tissue and letting it live in a dish for a short time. Today, most tissue culture experiments are done using cells rather than tissues. Much of what we know about cancer, heritable diseases, and the effects of the environment on human health has been derived from studies of cultured cells.
What types of cells do people study, and from where do they come? Cells acquired directly from animal tissue are called primary cells. They are difficult to culture, largely because primary cells in this context divide only a limited number of times. This limitation on the lifespan of cultured primary cells, called the Hayflick limit, is a problem because it requires a researcher to repeatedly remove tissues from animals to complete a study. Cell isolation processes can be quite labor-intensive, and also can complicate data analysis due to inherent animal-to-animal variation. To get around this problem, researchers study cells that are immortal, which means the cells are able to divide indefinitely. Though using immortal cells is preferable for many reasons, some inherent cell-to-cell variation still exists in such populations and the genetic changes that cause immortality may affect experimental outcomes.
The art of tissue culture lies in the ability to create conditions that are similar to what a cell would experience in an animal, namely 37°C and neutral pH. Blood nourishes the cells in an animal, and blood components are therefore used to feed cells in culture. Serum, the cell-free (and clotting-factor free) component of blood, contains many of the factors necessary to support the growth of cells outside the animal. Consequently, serum is frequently added to tissue culture medium, although serum-free media (also called chemically defined media) exist and support some types of cultured cells.
Cultured mammalian cells must grow in a germ-free environment and researchers using tissue culture must be skilled in sterile or aseptic technique. Bacteria double very quickly relative to mammalian cells. An average mammalian cell doubles about once per day whereas many common bacteria can double every 20 minutes under optimal conditions. Consequently, if you were to co-culture 100 mammalian cells and 1 bacteria together in a flask, within 24 hours you would have ~200 unhappy mammalian cells, and about 100 million happy bacteria! Needless to say, you would not find it very useful to continue to study the behavior of your mammalian cells under these conditions.
Today you will learn and practice aseptic technique to collect and count cells that were previously cultured by the teaching faculty. You will use the cells you collect to seed (or culture) cells for a survival assay that will assess the effect of drug treatment following DNA damage. To ensure you are well-prepared for your work in the tissue culture room, your instructor will provide a demonstration before you begin. Because the tissue culture room is not large enough to accommodate the entire class at once, we will be split into two groups today... and all subsequent days that involve tissue culture. Half of the class will begin in tissue culture with Part 1 and the other half of the class will begin by preparing previously seeded DLD-1 and BRCA2- cells for Western blot analysis. Your instructor will designate the groups.
Protocols
Part 1: Practice tissue culture techniques
As you prepare to work in the tissue culture room, review the following resource:
Part 1a: Demonstration of best practices
The teaching faculty will show you several key techniques that will be important to your success in tissue culture. Please carefully observe the following procedures, take notes, and ask questions!
- Prepare the tissue culture hood.
- Observe your cells.
- Trypsinization of cultured cells.
- Calculate cell culture density.
- Seed cells.
- Clean tissue culture hood.
Part 1b: Seed cells for RNA purification
You will use the techniques you learned during the demonstration to seed T75 flasks with cells. These cells will be used to assess the effect of DNA damage on gene expression. More specifically, you will examine this effect in the BRCA2- mutant cells compared to the DLD-1 wild-type cells.
Each team will receive two T75 culture flasks: one with DLD-1 (wild-type) cells, and one with BRCA2- mutant cells. You will enzymatically detach these adherent cells, count them, and seed them at specific densities in T75 flasks.
It is essential that you do not mix up or cross-contaminate the DLD-1 and BRCA2- cell lines! We suggest that one partner is responsible for each flask, and that partners do not share Pasteur pipets or other equipment.
Preparing tissue culture hood
- The tissue culture hood is partly set up for you. Finish preparing your hood according to the demonstration, first bringing in any remaining supplies you will need, then obtaining the pre-warmed reagents from the water bath, and finally retrieving your cells from the 37 °C incubator.
- Don't forget to spray everything down with 70% ethanol!
- One of the greatest sources for tissue culture contamination is moving materials in and out of the hood because this disturbs the air flow that maintains a sterile environment inside the hood. Think about what you will need during your experiment to avoid moving your arms in and out of the hood while you are handling your cells.
Collecting cells
- Examine your cell cultures as you remove the flasks from the incubator.
- Look first at the color and clarity of the media. Fresh media is reddish-orange in color and if the media in your flask is yellow or cloudy, it could mean that the cells are overgrown, contaminated, or starved for CO2.
- Next, look at the cells using the inverted microscope. Note their shape, arrangement, and how densely the cells cover the surface of the flask.
- After you look at your cells, take the flask to your tissue culture hood to begin the seeding procedure.
- Aspirate the media from the cells using a sterile Pasteur pipet.
- Wash the cells by adding 5 mL PBS using a 5 mL pipet. Slightly tip the flask back and forth to rinse the cells then aspirate the PBS with a fresh Pasteur pipet.
- To dislodge the cells from the flask, you will add trypsin, a proteolytic enzyme.
- With a 2 mL pipet, add 2 mL of trypsin to the flask.
- 2 mL pipets are tricky! They fill up quickly. Be careful not to pull up the liquid too quickly or it will go all the way up your pipet into the pipet-aid! If this happens, please alert the teaching faculty rather than returning the pipet-aid to the rack.
- Tip the flask in each direction to distribute the trypsin evenly then incubate the cells at 37°C for 10 minutes using a timer.
- This is a great time to clear out your trash and read ahead!
- Retrieve your flasks from the incubator and firmly tap the bottom 10 times to dislodge the cells.
- Check your cells using the microscope to ensure they are dislodged. They should appear round and move freely.
- If your cells are not detached from the flask, incubate at 37 °C for an additional minute.
- When your cells are dislodged, move your flask back into the tissue culture hood and add 5 mL of media to the cells then pipet the liquid up and down (“triturate”) to break up cells that are clumped together and suspend them in the liquid.
- Note: do not take up or release all the liquid, in order to avoid bubbles.
- Transfer the suspended cells into a labeled 15 mL conical tube
- Transfer 90 μL of your cell suspension from the 15 mL conical tube into a labeled eppendorf tube.
- Be sure to cap your conical tube and eppendorf tube after you transfer your cells.
Counting cells
- Remove the eppendorf with your cells from the tissue culture hood.
- Add 10 μL of Trypan blue to the eppendorf tube and pipet up and down to mix.
- With a new pipet tip, carefully pipet 10 μL of the stained cells between the hemocytometer and (weighted) glass cover slip, as shown to you by the teaching faculty.
- Keep BRCA2- on the 'top' and DLD-1 on the 'bottom', so you don't forget which is which.
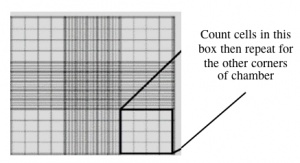
- Count the cells that fall within the four corner squares (with a 4x4 etched grid pattern), average (i.e. divide by 4), and then multiply by 10,000 to determine the number of cells/mL.
Seeding cells
- Obtain four T75 flasks and label two DLD-1 and two BRCA2-.
- Include your section information and the date.
- Calculate the volume of each cell suspension you need to seed 100,000 DLD-1 cells and 250,000 BRCA2- cells per T75 flask.
- For example, if your concentration is 1 million (1 M) cells/mL, you would seed the fresh T75 flasks 250 μL of DLD-1 cells.
- Subtract the volume of cell suspension that you calculated in Step #2 from 12 mL.
- Add this volume of media to the fresh T75 flask.
- Add the volume of cell suspension that you calculated in Step #2.
- You should mix your cell suspension by pipetting before distributing the small volume of cells to the flasks as the cells likely settled to the bottom of the conical while you were counting.
- Finally, tilt your flasks back and forth to distribute the cells evenly.
- Before moving your flasks to the 37 °C incubator, use the microscope to visually confirm that cells are present.
Cleaning the tissue culture hood
The next group who uses your hood should find the surfaces wiped down and free of equipment.
- Aspirate any remaining cell suspensions.
- Dispose of all vessels that held cells in the biohazard waste box and be sure that all sharps are in the sharps jar.
- Remove any equipment or supplies that you transferred into the hood and return to the appropriate location.
- Please leave the equipment that was already there.
- Spray the TC hood surface with 70% ethanol and wipe with paper towels.
- Be sure the paper towels are disposed of in the biohazard waste box!
- Empty the benchtop biohazard bucket into the biohazard waste box.
Part 2: Begin Western blot
You will use cells seeded by the teaching faculty to prepare cell lysates that will be used to confirm that BRCA2 is indeed absent from the mutant cell line, and present in the parental cell line.
Part 2a: Prepare cell lysates
In this exercise you will prepare your samples for Western blot analysis. It is very important that you keep the cell lysate cold throughout the procedure.
- You have an ice bucket at your bench with the following pre-chilled items inside: two empty eppendorfs, RIPA buffer, protease inhibitors, and PBS.
- Label the eppendorf tubes as DLD-1 and BRCA2- (plus your section information).
- Retrieve two 60 mm dishes from the small 37 °C incubator: one that contains DLD-1 cells and one that contains BRCA2- cells.
- Place the dishes at a 30-45° angle in your ice bucket.
- You may find it helpful to push the ice such that you build a ramp in the bucket that you can use to hold the plate steady.
- Add 2.5 μL of protease inhibitors to your 250 μL of aliquotted RIPA buffer and return this mixture to the ice.
- Aspirate the media from each well by holding the tip of the pasteur pipet (be sure to cap the pasteur pipet with a clean yellow tip!) at the bottom of the well.
- Add 2 mL of ice-cold PBS to each dish.
- Aspirate the ice-cold PBS – be sure to remove ALL of the PBS after this wash.
- Add 100 μL of RIPA lysis buffer to the top of each dish and allow it to run down the dish to the bottom.
- Dislodge the cells from the dish by scraping each well with a fresh cell scraper.
- First tilt the plate back and forth to coat the cells with lysis buffer.
- Then go from top to bottom 'windshield wiper style' to pool the cells at the bottom of each well in the tilted plate.
- Transfer the cells from each dish to the appropriate eppendorf tube.
- At this step it is important to make sure there are no aggregates or clumps of cell lysate remaining in the dish. Do this by tilting the plate so that light reflects off the bottom. If you see any 'chunks', pipette your lysate back into the dish to dissolve the chunk, then re-transfer everything into the eppendorf tube.
- Incubate the eppendorf tubes with your cell lysates on ice for 10 min.
- Toward the end of this incubation bring your tubes, on ice, to the front bench to sonicate the lysate for 10seconds, followed by 30 seconds on ice. Repeat this 3 times.
- Spin the cell lysate at maximum speed in the cold room centrifuge for 10 min to pellet insoluble material.
- The teaching faculty will take you to the centrifuge when you are ready.
- Meanwhile, label two fresh eppendorf tubes and add them to your ice bucket to chill for a later step.
- This step is typically referred to as "clearing" the lysate. The pellet at the bottom contains the DNA from the cell, and genomic DNA can get very soupy making it difficult to load your lysate on the SDS-PAGE gel.
- Transfer the supernatant to the fresh eppendorf tubes – be careful not to disturb the pellet at the bottom!
- Keep your samples on ice when not in use.
Part 2b: Measure protein concentration
You will now measure the total protein concentration in each cell lysate to determine the volume that you will use for the polyacrylamide gel separation.
- Prepare your assay reagent by mixing 1 part BioRad dye with 4 parts deionized water.
- You will need 1 mL of diluted dye per sample, this will be enough for your blank and two cell lysate samples.
- This reagent is hazardous chemical waste. Teaching staff will collect all used and unused samples once measurements are complete.
- Prepare a 'blank' by adding 1 mL of diluted dye then 1 μL of your leftover RIPA buffer to a cuvette. Cover the cuvette with parafild then invert to mix.
- Add 1 mL of diluted dye then 1 μL of each cell lysate to a plastic cuvette. As above, cover with parafilm and invert to mix.
- Incubate cuvettes with dye plus sample for a minimum of 5 min at room temperature.
- Samples should incubate at room temperature for no more than 1 h.
- Measure the absorbance of each sample at 595 nm.
- Use the RIPA sample to blank the spectrophotometer prior to measuring samples.
- Calculate the protein concentrations of your DLD-1 and BRCA2- cell lysate using the following information:
- Standard curve linear regression: y = 0.058*x - 0.018, where y is your measured absorbance and x is your sample protein concentration in μg/μL.
- Next, calculate the volumes of lysate and water required to add 40 μg of total protein to the polyacrylamide gel in a total volume of 20 μL, per lysate sample.
- If your concentration is greater than 2 μg/μL, use water to make up the remaining volume.
- If your concentration is less than 2 μg/μL for at least one sample, scale both samples down to a lower protein amount, such as 20 μg.
- Do not throw away the remainder of your cell lysate! We will store these samples at -80 °C in case your Western blot needs to be repeated.
Part 2c: Prepare cell lysates for polyacrylamide gel electrophoresis (PAGE)
- Add 4 μL of 6X Laemmli sample buffer to the cell lysate samples you prepared in Part 2b Step #7.
- Briefly vortex each tube, then centrifuge to collect the samples at the bottom of the tubes.
- Put lid locks on the eppendorf tubes and boil for 5 minutes in the water bath that is in the fume hood.
- Allow your samples to cool to room temperature, then give them to the teaching faculty for storage at -20 °C until your next laboratory session.
Part 3: Review research paper
We will end today with a discussion of the Dietlein et al. research article. In their research, the authors completed a screen to examine 1319 cancer-associated genes from 67 cell lines to identify cancer-cell specific mutations that are associated with DNA-PKcs dependence or addiction. Refer to the M2 overview for more information on addiction in cancer cells.
Our paper discussion will assist you in writing a cohesive story that clearly reports the data and provides strong support for the conclusions made about the data. During the paper discussion, everyone is expected to participate - either by volunteering or by being called upon!
Introduction
Remember the key components of an introduction:
- What is the big picture?
- Is the importance of this research clear?
- Are you provided with the information you need to understand the research?
- Do the authors include a preview of the key results?
Results
Carefully examine the figures. First, read the captions and use the information to 'interpret' the data presented within the image. Second, read the text within the results section that describes the figure.
- Do you agree with the conclusion(s) reached by the authors?
- What controls are included and are they appropriate for the experiment performed?
- Are you convinced that the data are accurate and/or representative?
Discussion
Consider the following components of a discussion:
- Are the results summarized?
- Did the authors 'tie' the data together into a cohesive and well-interpreted story?
- Do the authors overreach when interpreting the data?
- Are the data linked back to the big picture from the introduction?
Reagents
From Thermo Fischer
- DLD-1 wild-type cells
- BRCA2-/- mutant cells
Media components from Life Technologies, Inc. unless noted otherwise.
- DLD-1 and BRCA2-/- cell media
- RPMI 1640
- 10% fetal bovine serum (FBS)
- 100X antibiotic solution from Cellgro
- 10,000 U/mL Penicillin
- 10,000 U/mL Streptomycin
- phosphate-buffered saline (PBS)
- 0.05% trypsin/0.91 mM EDTA
- Incubator maintains 37°C, 5% CO2 and 95% relative humidity
From Boston Bioproducts:
- RIPA lysis buffer
- 50 mM Tris-HCl, pH 7.4
- 150 mM NaCl
- 1% NP-40
- 0.5% sodium deoxycholate
- 0.1% SDS
- 100X protease inhibitor cocktail
- AEBSF
- Aprotinin
- E-64 Besstain Leupeptin
- EDTA
- 6x reducing Laemmli sample buffer
- 375 mM Tris HCl, pH 6.8
- 9% SDS
- 50% glycerol
- 9% betamercaptoethanol
- 0.03% bromophenol blue
From Bio-Rad
- Protein Assay Dye (5X)
Next day: Prepare Western blot and induce DNA damage for quantitative PCR assay
Previous day: Last day of M1