20.109(S21):M3D2
Contents
Introduction
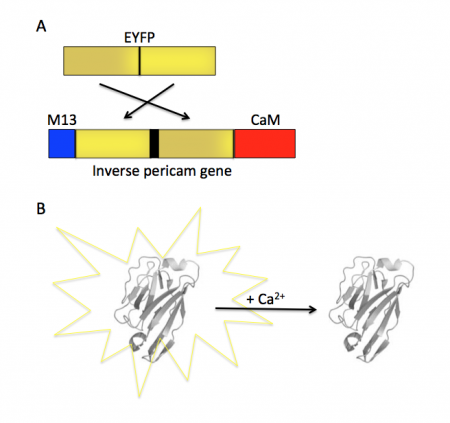
Today you will familiarize yourself with the recombinant protein IPC and its constituent parts. The fluorescent component of IPC is an enhanced yellow fluorescent protein (abbreviated EYFP), one of the many derivatives of green fluorescent protein (GFP). GFP is naturally produced by jellyfish and was cloned into other organisms in the early 1990’s. It has since been exploited as a genetically encodable reporter and mutagenized to vary its excitation and emission spectra. The other key component of inverse pericam is the protein calmodulin (CaM), a natural calcium sensor that is present in all eukaryotes Calmodulin has many ligands that it binds only in the presence of calcium ion, including the peptide fragment M13. This conditional specificity for M13 binding is enabled by the change in confirmation of CaM when bound to calcium.
Within inverse pericam, M13 and CaM are located at opposite ends, surrounding a permuted (i.e., rearranged) version of EYFP. In the absence of calcium, this EYFP exhibits strong fluorescence. However, when enough calcium is added to a solution of inverse pericam, CaM and M13 interact, disrupting the conformation and, as a result, the fluorescence of EYFP. The transition from bright to dim fluorescence occurs over a particular concentration range of calcium. The calcium concentration at which binding to CaM occurs (and fluorescence decreases) is referred to as the Kd and determined by the affinity of CaM to calcium. In addition, the interaction between CaM and calcium is impacted by cooperativity. CaM has four calcium binding sites. In cooperativity, the affinity of CaM for calcium is altered by how many calcium ions are already bound to the protein. The mutations you will examine today were designed in an effort to modify the calcium sensor portion of IPC in a manner that is likely to change the affinity and / or cooperativity for calcium ions.
To examine the modification that were made to IPC, we will use several protein analysis tools. Proteins are modular materials that may be described and examined at multiple levels of a structural hierarchy (from primary to quaternary in the classical paradigm). Primary structure refers to a protein’s amino acid sequence, which might reveal a cluster of charged residues or a pattern of alternating polar and nonpolar residues. One cannot predict off-hand the conformation of a protein merely from its linear sequence; however, due to rotational flexibility of bonds and non-covalent interactions between non-adjacent amino acids (as well as covalent disulfide bonds) some structural characteristics can be inferred. Because many proteins have structural motifs in common (e.g., alpha helices and beta sheets at the secondary level, or leucine-rich repeats at the tertiary level), which ultimately arise from the amino acid sequences, databases can be useful for making predictions about proteins with known amino acid sequences but unknown structures.
Protocols
Part 1: Review IPC sequence and structure
Part 2: Examine IPC mutations
Reagents list
Next day: Induce and purify IPC variants