20.109(F20):M3D5
Part 3: Design gRNA for CRISPRi system
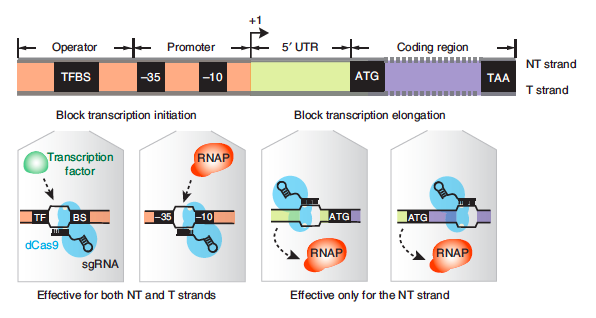
Now that you know which gene you will target with the CRIPSRi system, you need to design a guide RNA (gRNA) using the DNA sequence. As a reminder, the gRNA recruits dCas9 to a specific location in the genome where it will bind and block transcription. The CRISPRi system can block transcription at either the initiation or elongation steps of transcription (see below image). If the dCas9 protein is bound to the promoter sequence at either a transcription factor binding site (TFBS) or the -10/-35 boxes it will block transcription factors and/or RNA polymerase from binding to the promoter of the gene, thereby impeding transcription initiation. If the dCas9 protein is bound within the gene it impede RNA polymerase elongation of the transcript. Consider both options with your partner and decide on a strategy before working through the remaining protocol. And, as always, record your thoughts in your notebook.
When you decide on which region of your target gene you will use to design your gRNA follow the steps below to choose a gRNA sequence.
- Use the KEGG Database to obtain the DNA sequence of your target gene in the E. coli K-12 MG1655 strain.
- Enter the name of your target gene in the Search genes box and click Go.
- Double click on the linked gene name.
- Using the information provided by the KEGG database, answer the following questions:
- What is the full name of your gene (or Definition)?
- In what pathways is your gene involved?
- Determine the position of your gene within the genome by clicking the Genome map button within the Position row.
- Is your target gene at the start of an operon or in the middle?
- Does the position of your gene within the operon affect the gRNA strategy you discussed with your partner? If you are unsure, consult with the teaching faculty.
- The amino acid (AA) sequence and nucleotide sequence (NT) of your gene are provided at the bottom of the page.
- Generate a new DNA file in SnapGene that contains the NT sequence of your gene.
- If you will design a gRNA that binds the promoter, enter 50 in the +upstream box to get the 50 basepair sequence immediately preceding the start codon.
- Use the DNA sequence to select a 20-25 basepair region that will serve as the gRNA binding site. Please make sure your sequence does not exceed 25 bp.
- If you target the template DNA strand, the gRNA sequence will be the same as the transcribed sequence.
- If you target the nontemplate strand, the gRNA sequence will be the reverse-complement of the transcribed sequence.
- To avoid off-target effects, check the specificity of your gRNA sequence using the Basic Local Alignment Search Tool (BLAST) available from NCBI.
- Click on the Nucleotide BLAST icon.
- In the Enter Query Sequence section, paste your gRNA sequence into the Enter accession number(s), gi(s), or FASTA sequence(s) box.
- In the Choose Search Set section, type 'MG1655' in the Organism box and select an appropriate option from the drop down list.
- Click the BLAST button at the bottom of the screen.
- Ideally, your gRNA sequence will only have 100% identity to the target sequence.
- If your gRNA sequence binds to multiple locations (with a 14+ nucleotide region within the gRNA having 100% identity to an off-target) within the genome, consider redesigning your gRNA.
- When you are satisfied with your gRNA sequence selection, post it to the Class Data Page.
- Be sure you write your gRNA sequence 5' → 3'.
Additional sequence information must be added to your gRNA for the CRISPRi system. As discussed in lecture, a 'tag' is required for dCas9 binding at the target site . The tag sequence 5' - GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC - 3' will be included at the 3' end of your gRNA. Please double check that in total, your full sequence is 60 bp or fewer.
The teaching faculty will order your gRNA as an oligo or primer from Genewiz.