20.109(F20):M2D2
Introduction
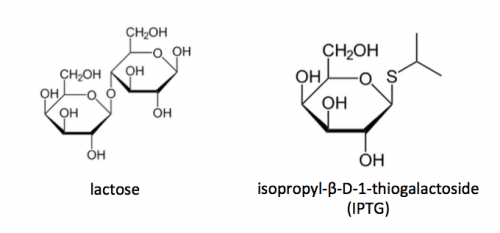
To induce production of PF3D7_1351100 protein from the expression plasmid that was 'cloned' in the previous laboratory session, a lactose-analogue isopropyl β-D-1-thiogalactopyranoside (IPTG) and arabinose was used to induce expression in Nico(DE3) E. coli bacterial cells. The use of IPTG to induce protein expression is based on the native lac operon used for lactose metabolism in bacterial cells.
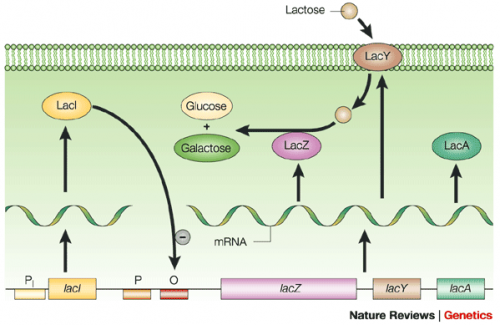
The lac operon is composed of four genes: lacI, lacZ, lacY, and lacA. When lactose is absent, LacI (the protein encoded by lacI) binds to the operator sequence (O) upstream of lacZYA. In the presence of lactose, LacI and lactose form a complex which relieves repression of lacZYA transcription. LacZ is a β-galactosidase that cleaves lactose resulting in glucose and galactose. LacY, a β-galactoside permease, facilitates the transport of lactose across the cell membrane, and LacA, a β-galactoside transacetylase, transfers an acetyl group from acetyl-CoA to β-galactosides.The native lac operon is a powerful tool in engineering protein expression systems because it enables researchers to control gene expression using inducer molecules. The lacZYA genes are only expressed when lactose is present. If a gene of interest is cloned downstream of the operator sequence, the expression of this gene can be controlled by LacI repression and lactose derepression. To further control the system for protein expression, IPTG is used as a lactose-analog as it is not metabolized by the cells.
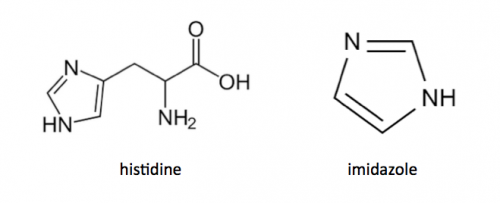
Today you will isolate the expressed PF3D7_1351100 protein from the bacterial cells. Remember that the PF3D7_1351100 gene sequence was synthesized using a gBlock and 6xHis-tag was added to the DNA sequence. The resultant protein is therefore His-tagged. Histidine has several interesting properties, notably its near-neutral pKa, and His-rich peptides are promiscuous binders, particularly to metals. (For example, histidine side chains help coordinate iron molecules in hemoglobin.)
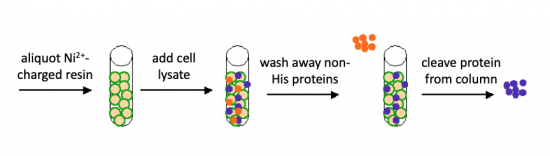
You will use a nickel-agarose resin to separate PF3D7_1351100 from the other proteins present in the bacteria. The His-tagged protein will preferentially bind to the nickel-coated beads, while proteins irrelevant to our purposes in Module 1 can be washed away using imidazole. Remember, the BL21-A1 cells are not only producing your protein, but also the proteins needed for cellular function and survival. Because imidazole is also able to bind to nickel, washing the column with a low concentration solution will promote competition for binding between the imidazole and bound proteins. Proteins that are non-specifically bound will have a lower affinity for the nickel than imidazole and be washed from the column, whereas the 6xHis-tagged TDP43-RRM12 will remain adhered to the nickel-agarose resin. Finally, an enzyme cleavage reaction will be used to elute TDP43-RRM12 from the nickel-agarose resin.