Difference between revisions of "20.109(S21):M3D2"
Noreen Lyell (Talk | contribs) (→Reagents list) |
Noreen Lyell (Talk | contribs) (→Part 1: Identify mutations in Variant IPC proteins) |
||
| Line 17: | Line 17: | ||
===Part 1: Identify mutations in Variant IPC proteins=== | ===Part 1: Identify mutations in Variant IPC proteins=== | ||
| + | In an effort to gain further insight into calcium affinity / cooperativity of the IPC calcium sensor, you will analyze the data generated from six Variant IPC proteins. The first step in this process is to identify the mutations that were incorporated into the Variant IPC sequences. To do this you will perform sequence alignments (just as you did on M1D5!). | ||
| + | |||
| + | # | ||
===Part 2: Review expression system used to express IPC and IPC variants=== | ===Part 2: Review expression system used to express IPC and IPC variants=== | ||
Revision as of 12:41, 20 April 2021
Contents
Introduction
In the previous laboratory session, you examined the structural features of IPC. The goal of this was to consider which residues are important to affinity and / or cooperativity. Today you will consider the mutations that were generated in IPC by previous 109ers and review how IPC and the IPC variants were expressed and purified as this is the first step in testing how the variants perform as calcium sensors.
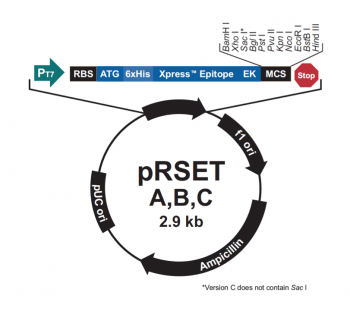
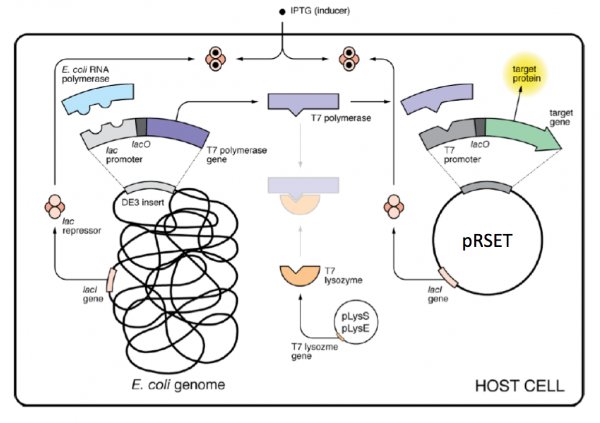
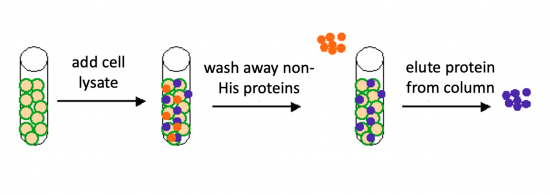
The genetic sequences that encode the IPC protein and IPC variant proteins are maintained within the pRSET expression vector (recall the cloning exercise from M3D1!). This expression vector contains several features that are important to the expression and purification of IPC and the IPC variants. To enable selection of bacterial cells that carry pRSET_IPC, an antibiotic cassette, specifically an ampicillin marker, is included on the vector. The features most relevant to protein expression and purification are highlighted in the schematic to the right. The T7 promoter drives expression of the gene that encodes IPC (or IPC variant). To ensure that the transcript is translated into a protein, a ribosome binding site (RBS) is included. The ATG sequence serves as the transcriptional start and the 6xHis represents the six-histidine residue tag that is used for protein purification via affinity chromatography.There are some similarities between the expression system used to purify TDP43-RRM12 in Mod 2 and the system we will use for IPC. First, in both expression systems IPTG is used to induce protein production. As a review, IPTG is a lactose analog that induces expression by binding to the LacI repressor. When bound to IPTG, the LacI repressor is not able to bind to the lac operator sequence and transcription occurs unimpeded. For more details please review to the M2D1 Introduction! Another similarity is that a 6His tag is used and 6His-tagged IPC and IPC variants will purified using column affinity as shown in the image below. There are also several differences between the expression systems for TDP43-RRM12 and IPC. As you read through the exercises below, consider how these steps are different from those used previously.
Protocols
Part 1: Identify mutations in Variant IPC proteins
In an effort to gain further insight into calcium affinity / cooperativity of the IPC calcium sensor, you will analyze the data generated from six Variant IPC proteins. The first step in this process is to identify the mutations that were incorporated into the Variant IPC sequences. To do this you will perform sequence alignments (just as you did on M1D5!).
Part 2: Review expression system used to express IPC and IPC variants
As mentioned above, IPTG is used to induce protein production in the expression systems for TDP43-RRM12 and IPC; however, the mechanism that drives transcription of the gene that encodes the protein of interest is different. For IPC and the IPC variants, the proteins are expressed using the BL21(DE3)pLysS strain of E. coli, which has the following genotype: F-, ompT hsdSB (rB- mB-) gal dcm (DE3) pLysS (CamR).
The expression vector, pRSET, encodes the bacteriophage T7 promoter, which is active only in the presence of T7 RNA polymerase (T7RNAP), an enzyme that therefore must be expressed by the bacterial strain used to make the protein of interest. In BL21(DE3), T7RNAP is associated with a lac construct. Constitutively expressed lac repressor (lacI gene) blocks expression from the lac promoter; thus, the polymerase will not be produced except in the presence of repressor-binding lactose or a small-molecule lactose analogue such as IPTG (isopropyl β-D-thiogalactoside). To reduce ‘leaky’ expression of the protein of interest (in our case, IPC), the pLysS version of BL21(DE3) contains T7 lysozyme, which inhibits basal transcription of T7RNAP. This gene is retained by chloramphenicol selection, while the pRSET plasmid itself (and thus IPC) is retained by ampicillin selection.
The pRSET_IPC and pRSET_IPC variants were transformed into chemically competent BL21(DE3)pLysS using heat shock as described previously. To review this method, look back at the information provided on M1D3!
In your laboratory notebook, complete the following:
- questions about expression system...
Part 3: Evaluate purified IPC
Quick note on cloning and purification...
To evaluate the purified IPC protein, we will use the same methods as when we assessed purified TDP43-RRM12: SDS-PAGE and microBCA (this is a variation of the BCA procedure that is used to measure lower protein concentrations). To review these methods, look back at the information provided on M2D2!
Assess purity using SDS-PAGE
In your laboratory notebook, complete the following:
- Evaluate the purified protein product for each of the Variant IPC and the WT IPC.
- Do you see a band that corresponds to the expected protein size? Edit the image of the SDS-PAGE results such that the expected band in each lane is highlighted.
- Attach the edited image.
- Are the purified protein products for each of the Variant IPC and the WT IPC pure?
- Estimate how much of the total protein in each purified protein product is the Variant IPC or WT IPC. (Note: this is just an estimate, do your best to gauge the percentage of Variant IPC or WT IPC protein in each sample by considering the intensities of all of the bands in the lane.)
Measure concentration using microBCA
In your laboratory notebook, complete the following:
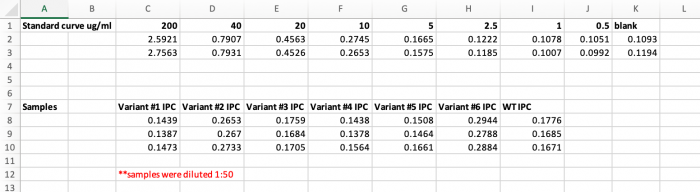
- Use the microBCA data to generate a standard curve and calculate the protein concentrations for the Variant IPC and WT IPC purification samples (spreadsheet with values attached here).
- Attach the standard curve and the calculations for the Variant IPC and WT IPC concentration calculations.
- Based on the percentage you estimated above, what is the actual concentration of the Variant IPC and WT IPC in the purification samples.
Next day: Evaluate effect of mutations on IPC variants