Difference between revisions of "20.109(S20):Purify RNA from etoposide-treated cells and and generate cDNA (Day3)"
Noreen Lyell (Talk | contribs) (→Protocols) |
Noreen Lyell (Talk | contribs) (→Protocols) |
||
| (5 intermediate revisions by one user not shown) | |||
| Line 6: | Line 6: | ||
Differential gene expression is used to translate information perceived by a cell into a gene product. All forms of life rely on gene expression to generate proteins and non-protein coding gene products that provide the machinery for cell survival. In this, gene expression is the most basic mechanism by which genotype results in a phenotype. One method researchers use to examine gene expression is to quantify transcript levels for specific genes. | Differential gene expression is used to translate information perceived by a cell into a gene product. All forms of life rely on gene expression to generate proteins and non-protein coding gene products that provide the machinery for cell survival. In this, gene expression is the most basic mechanism by which genotype results in a phenotype. One method researchers use to examine gene expression is to quantify transcript levels for specific genes. | ||
| − | In this module, we are interested in the effect of | + | In this module, we are interested in the effect of etoposide on gene expression. To address these questions we are using the DLD-1 cell line. In chemotherapy, drugs that induce DNA damage are used to specifically target cancer cells. Due to the faster growth rate of cancer cells, DNA damaging drugs are able to impact survival moreso than in healthy cells. |
We will use two experimental approaches to evaluate gene expression: qPCR and RNA-seq. Though the intricacies of each method will be discussed later, a critical first step for both is the purification of high-quality RNA. RNA purification is complicated by ribonuclease enzymes, such as RNase. RNases are ubiquitous can be difficult to neutralize. Furthermore, RNases are present in the cells and tissues from which the RNA is isolated. The 'gold standard' for RNA purification is an organic extraction method. In this process a sample is homogenized in a phenol solution then centrifuged, which separates the sample into three phases: a lower organic layer, a denatured protein and gDNA middle layer, and an RNA upper layer. The RNA from the upper layer is alcohol precipitated and rehydrated. Because this method is labor-intensive and requires chlorinated organic reagents, researchers often use column-based purification kits. | We will use two experimental approaches to evaluate gene expression: qPCR and RNA-seq. Though the intricacies of each method will be discussed later, a critical first step for both is the purification of high-quality RNA. RNA purification is complicated by ribonuclease enzymes, such as RNase. RNases are ubiquitous can be difficult to neutralize. Furthermore, RNases are present in the cells and tissues from which the RNA is isolated. The 'gold standard' for RNA purification is an organic extraction method. In this process a sample is homogenized in a phenol solution then centrifuged, which separates the sample into three phases: a lower organic layer, a denatured protein and gDNA middle layer, and an RNA upper layer. The RNA from the upper layer is alcohol precipitated and rehydrated. Because this method is labor-intensive and requires chlorinated organic reagents, researchers often use column-based purification kits. | ||
| Line 17: | Line 17: | ||
==Protocols== | ==Protocols== | ||
| − | |||
| − | |||
===Part 1: Prepare samples for quantitative PCR assay=== | ===Part 1: Prepare samples for quantitative PCR assay=== | ||
| Line 24: | Line 22: | ||
First, spray and wipe down your bench with 70% ethanol. Then, obtain a piece of absorbent paper. This will be your RNAse free work space. Spray the absorbent paper with RNase Away and dry with a Kimwipe. Perform this cleaning procedure with all of the equipment you will need for the RNA purification protocol (read through Part 2a Step #11 to the end of the protocol). Also, obtain all of the aliquots you will need from the front laboratory bench and clean before placing the tubes on the absorbent paper work space. Lastly, obtain a 50 mL conical tube and label it 'waste'. | First, spray and wipe down your bench with 70% ethanol. Then, obtain a piece of absorbent paper. This will be your RNAse free work space. Spray the absorbent paper with RNase Away and dry with a Kimwipe. Perform this cleaning procedure with all of the equipment you will need for the RNA purification protocol (read through Part 2a Step #11 to the end of the protocol). Also, obtain all of the aliquots you will need from the front laboratory bench and clean before placing the tubes on the absorbent paper work space. Lastly, obtain a 50 mL conical tube and label it 'waste'. | ||
| − | |||
| − | |||
====Part 1a: Purify RNA from cells==== | ====Part 1a: Purify RNA from cells==== | ||
#Prepare your working space in the tissue culture hood. | #Prepare your working space in the tissue culture hood. | ||
| − | #Retrieve your | + | #Retrieve your T75 flask (DLD-1 +etop) from the 37 °C incubator and visually inspect your cells with a microscope. |
| + | #*Remember, the instructors prepared the RNA that you will use for comparison to +etop treatment. | ||
#*Record your observations concerning media color, confluency, etc. in your laboratory notebook. | #*Record your observations concerning media color, confluency, etc. in your laboratory notebook. | ||
| − | #*Cells were seeded | + | #*Cells were seeded such that ~1x10^6 cells are harvested for RNA purification. |
#In the tissue culture room, aspirate the media from each flask. | #In the tissue culture room, aspirate the media from each flask. | ||
#Wash the cells by adding 3 mL PBS using a 5 mL pipet. Slightly tip the flask back and forth to rinse the cells then aspirate the PBS by covering the pasteur pipet attached to the aspirator with a yellow tip. | #Wash the cells by adding 3 mL PBS using a 5 mL pipet. Slightly tip the flask back and forth to rinse the cells then aspirate the PBS by covering the pasteur pipet attached to the aspirator with a yellow tip. | ||
| − | |||
#With a 2 mL pipet, add 2 mL of trypsin to each flask. | #With a 2 mL pipet, add 2 mL of trypsin to each flask. | ||
#Tip the flask in each direction to distribute the trypsin evenly then incubate the cells at 37 °C incubator for 5 minutes using a timer. | #Tip the flask in each direction to distribute the trypsin evenly then incubate the cells at 37 °C incubator for 5 minutes using a timer. | ||
| − | #Retrieve your | + | #Retrieve your flask from the incubator and firmly tap the bottom 10 times to dislodge the cells. |
#*Check your cells using the microscope to ensure they are dislodged. They should appear round and move freely. | #*Check your cells using the microscope to ensure they are dislodged. They should appear round and move freely. | ||
#*If your cells are not detached from the flask, incubate at 37 °C for an additional minute. | #*If your cells are not detached from the flask, incubate at 37 °C for an additional minute. | ||
| − | #When your cells are dislodged, add 3 mL of PBS to | + | #When your cells are dislodged, add 3 mL of PBS to the flask. |
#*To ensure you collect all of the cells in your flask, pipet the PBS down the bottom of the flask to wash the cells from the surface. | #*To ensure you collect all of the cells in your flask, pipet the PBS down the bottom of the flask to wash the cells from the surface. | ||
#*Pipet up and down 3 times. | #*Pipet up and down 3 times. | ||
#*Note: do not take up or release all the liquid, in order to avoid bubbles. | #*Note: do not take up or release all the liquid, in order to avoid bubbles. | ||
#Transfer the suspended cells into a labeled 15 mL conical tube. | #Transfer the suspended cells into a labeled 15 mL conical tube. | ||
| − | #Centrifuge your | + | #Centrifuge your cell suspension at 1000 rpm for 5 min to pellet the cells. |
| − | #Carefully remove your | + | #Carefully remove your tube from the centrifuge and return to your laboratory bench in the main lab. |
| − | #*Avoid agitating your | + | #*Avoid agitating your tube as the pellet is easily disrupted. |
#Using a P1000 pipet, remove the supernatant from the tube into the 50 mL waste conical tube. | #Using a P1000 pipet, remove the supernatant from the tube into the 50 mL waste conical tube. | ||
#*'''Do not''' shake, tap, or flick the tubes to remove excess liquid as this will disrupt your cell pellet. | #*'''Do not''' shake, tap, or flick the tubes to remove excess liquid as this will disrupt your cell pellet. | ||
| Line 55: | Line 51: | ||
#*Only 700 μL at a time can be loaded onto the column. If you have more than 700 μL, consult the teaching faculty. | #*Only 700 μL at a time can be loaded onto the column. If you have more than 700 μL, consult the teaching faculty. | ||
#Centrifuge at 16,000 rpm for 2 min. | #Centrifuge at 16,000 rpm for 2 min. | ||
| − | #Remove the | + | #Remove the column from the collection tube and add 350 μL of 70% ethanol to the fluid remaining in the tube, then pipet to mix. |
#Transfer 700 μL of the RNA / ethanol suspension to an RNAeasy column (<font color='pink'>'''pink'''</font color>). | #Transfer 700 μL of the RNA / ethanol suspension to an RNAeasy column (<font color='pink'>'''pink'''</font color>). | ||
#Centrifuge at 10,000 rpm for 15 sec then discard the flow-through from the collection tube in your 50 mL waste conical tube. | #Centrifuge at 10,000 rpm for 15 sec then discard the flow-through from the collection tube in your 50 mL waste conical tube. | ||
| Line 64: | Line 60: | ||
#Move the columns into new collection tubes, then centrifuge at 16,000 rpm for 1 min. | #Move the columns into new collection tubes, then centrifuge at 16,000 rpm for 1 min. | ||
#Move the columns into 1.5 mL tubes (from the Qiagen RNeasy Mini Kit). | #Move the columns into 1.5 mL tubes (from the Qiagen RNeasy Mini Kit). | ||
| − | #*Label the base of the | + | #*Label the base of the tube to ensure you keep track of your samples. |
#Add 30 μL of RNase free water, then centrifuge at 10,000 rpm for 1 min. | #Add 30 μL of RNase free water, then centrifuge at 10,000 rpm for 1 min. | ||
#Alert the teaching faculty when you are done and you will be escorted to the NanoDrop to check the concentrations of your purified RNA samples. | #Alert the teaching faculty when you are done and you will be escorted to the NanoDrop to check the concentrations of your purified RNA samples. | ||
| Line 78: | Line 74: | ||
#Incubate the RNA-primer mixture in the 65 °C water bath on the front laboratory bench for 5 min. | #Incubate the RNA-primer mixture in the 65 °C water bath on the front laboratory bench for 5 min. | ||
#Retrieve your tube and incubate on ice for 1 min. | #Retrieve your tube and incubate on ice for 1 min. | ||
| − | #Prepare enough 'cDNA Synthesis Master Mix' for | + | #Prepare enough 'cDNA Synthesis Master Mix' for 2.1 reactions given the volumes of each component required for one reaction that are listed below. You will pipet the stock reagents at the front laboratory bench using filtered pipet tips. |
#*2 μL of 10X RT buffer | #*2 μL of 10X RT buffer | ||
#*4 μL of 25 mM MgCl<sub>2</sub> | #*4 μL of 25 mM MgCl<sub>2</sub> | ||
| Line 88: | Line 84: | ||
#Retrieve your tubes, leave on ice for 1min and add 1 μL of RNase H. Incubate in the 37 °C incubator for 20 min. | #Retrieve your tubes, leave on ice for 1min and add 1 μL of RNase H. Incubate in the 37 °C incubator for 20 min. | ||
#Your cDNA will be stored in the -20 °C freezer until the next laboratory session. | #Your cDNA will be stored in the -20 °C freezer until the next laboratory session. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Reagents== | ==Reagents== | ||
Latest revision as of 20:59, 28 February 2020
Contents
Introduction
Differential gene expression is used to translate information perceived by a cell into a gene product. All forms of life rely on gene expression to generate proteins and non-protein coding gene products that provide the machinery for cell survival. In this, gene expression is the most basic mechanism by which genotype results in a phenotype. One method researchers use to examine gene expression is to quantify transcript levels for specific genes.
In this module, we are interested in the effect of etoposide on gene expression. To address these questions we are using the DLD-1 cell line. In chemotherapy, drugs that induce DNA damage are used to specifically target cancer cells. Due to the faster growth rate of cancer cells, DNA damaging drugs are able to impact survival moreso than in healthy cells.
We will use two experimental approaches to evaluate gene expression: qPCR and RNA-seq. Though the intricacies of each method will be discussed later, a critical first step for both is the purification of high-quality RNA. RNA purification is complicated by ribonuclease enzymes, such as RNase. RNases are ubiquitous can be difficult to neutralize. Furthermore, RNases are present in the cells and tissues from which the RNA is isolated. The 'gold standard' for RNA purification is an organic extraction method. In this process a sample is homogenized in a phenol solution then centrifuged, which separates the sample into three phases: a lower organic layer, a denatured protein and gDNA middle layer, and an RNA upper layer. The RNA from the upper layer is alcohol precipitated and rehydrated. Because this method is labor-intensive and requires chlorinated organic reagents, researchers often use column-based purification kits.
We will use a commercially available column-based RNA purification system. Though the reagents are proprietary, the manufacturer provides some details concerning the inner workings of the kit. The key component is a silica-based membrane that binds RNA molecules longer that 200 nucleotides, which enriches for messenger RNA (mRNA) molecules. Prior to RNA isolation using the membrane, cells are lysed in the presence of a guanidine-thiocyanate-containing buffer that inactivates the RNases naturally present in the sample. Then ethanol is added to the lysate to promote binding of the RNA to the silica.
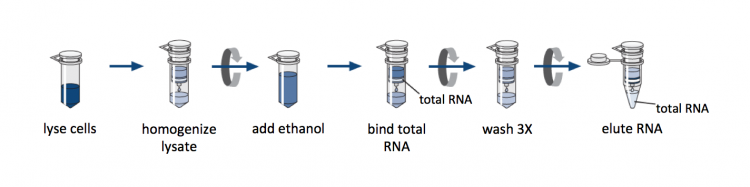
Following RNA purification, you will generate cDNA to use in your quantitative PCR analysis. Complementary DNA, or cDNA, refers to DNA that is generated from RNA. The ability to natively generate cDNA is unique to retroviruses. In research, this technology is available as a tool because the reverse transcriptase enzyme can be purified and used in exogenous reactions. We will generate cDNA from purified RNA to study gene expression as DNA molecules are more stable and, therefore, easier to work with experimentally. As with the RNA purification, we will use a kit to generate cDNA. The kit includes the necessary buffers and reverse transcriptase enzyme. First, the RNA is combined with a poly-T primer and oligonucleotides, then incubated at a high temperature for a denaturation step. An RNase inhibitor, reverse transcriptase, and buffers are then added. This mix is incubated at a temperature optimal for the reverse transcriptase enzyme to generate cDNA. The specific enzyme we will use is SuperScript III RT, and like the buffers included in the RNA purification kit, the genetic manipulations used to modify this reverse transcriptase enzyme are proprietary.
Protocols
Part 1: Prepare samples for quantitative PCR assay
You will move your cells to the main laboratory to purify the RNA from your cells. Before you harvest your cells in tissue culture, prepare your laboratory bench for work with RNA. As noted in the introduction, RNA is very sensitive to degradation and caution must be taken to preserve your sample.
First, spray and wipe down your bench with 70% ethanol. Then, obtain a piece of absorbent paper. This will be your RNAse free work space. Spray the absorbent paper with RNase Away and dry with a Kimwipe. Perform this cleaning procedure with all of the equipment you will need for the RNA purification protocol (read through Part 2a Step #11 to the end of the protocol). Also, obtain all of the aliquots you will need from the front laboratory bench and clean before placing the tubes on the absorbent paper work space. Lastly, obtain a 50 mL conical tube and label it 'waste'.
Part 1a: Purify RNA from cells
- Prepare your working space in the tissue culture hood.
- Retrieve your T75 flask (DLD-1 +etop) from the 37 °C incubator and visually inspect your cells with a microscope.
- Remember, the instructors prepared the RNA that you will use for comparison to +etop treatment.
- Record your observations concerning media color, confluency, etc. in your laboratory notebook.
- Cells were seeded such that ~1x10^6 cells are harvested for RNA purification.
- In the tissue culture room, aspirate the media from each flask.
- Wash the cells by adding 3 mL PBS using a 5 mL pipet. Slightly tip the flask back and forth to rinse the cells then aspirate the PBS by covering the pasteur pipet attached to the aspirator with a yellow tip.
- With a 2 mL pipet, add 2 mL of trypsin to each flask.
- Tip the flask in each direction to distribute the trypsin evenly then incubate the cells at 37 °C incubator for 5 minutes using a timer.
- Retrieve your flask from the incubator and firmly tap the bottom 10 times to dislodge the cells.
- Check your cells using the microscope to ensure they are dislodged. They should appear round and move freely.
- If your cells are not detached from the flask, incubate at 37 °C for an additional minute.
- When your cells are dislodged, add 3 mL of PBS to the flask.
- To ensure you collect all of the cells in your flask, pipet the PBS down the bottom of the flask to wash the cells from the surface.
- Pipet up and down 3 times.
- Note: do not take up or release all the liquid, in order to avoid bubbles.
- Transfer the suspended cells into a labeled 15 mL conical tube.
- Centrifuge your cell suspension at 1000 rpm for 5 min to pellet the cells.
- Carefully remove your tube from the centrifuge and return to your laboratory bench in the main lab.
- Avoid agitating your tube as the pellet is easily disrupted.
- Using a P1000 pipet, remove the supernatant from the tube into the 50 mL waste conical tube.
- Do not shake, tap, or flick the tubes to remove excess liquid as this will disrupt your cell pellet.
- Add 350 μL of RLT and pipet up and down to mix.
- Transfer the cell / RLT suspension to a Qiashredder column (purple).
- Be sure to label both the column insert and collection tube.
- Only 700 μL at a time can be loaded onto the column. If you have more than 700 μL, consult the teaching faculty.
- Centrifuge at 16,000 rpm for 2 min.
- Remove the column from the collection tube and add 350 μL of 70% ethanol to the fluid remaining in the tube, then pipet to mix.
- Transfer 700 μL of the RNA / ethanol suspension to an RNAeasy column (pink).
- Centrifuge at 10,000 rpm for 15 sec then discard the flow-through from the collection tube in your 50 mL waste conical tube.
- Repeat Steps #17 - 18 until all of the RNA / ethanol suspension has been passed through the RNAeasy column.
- Add 700 μL of RW1 and centrifuge at 10,000 rpm for 30 sec, then discard the flow-through in your 50 mL waste conical tube.
- Add 500 μL of RPE and centrifuge at 10,000 rpm for 30 sec, then discard the flow-through in your 50 mL waste conical tube.
- Add 500 μL of RPE and centrifuge at 10,000 rpm for 2 min, then discard the flow-through in your 50 mL waste conical tube.
- Move the columns into new collection tubes, then centrifuge at 16,000 rpm for 1 min.
- Move the columns into 1.5 mL tubes (from the Qiagen RNeasy Mini Kit).
- Label the base of the tube to ensure you keep track of your samples.
- Add 30 μL of RNase free water, then centrifuge at 10,000 rpm for 1 min.
- Alert the teaching faculty when you are done and you will be escorted to the NanoDrop to check the concentrations of your purified RNA samples.
Return your 50 mL 'waste' conical tube to the front bench. It will be disposed of as hazardous waste.
Part 1b: Generate cDNA
- Calculate the volume of your RNA solution that contains 1 μg of RNA.
- Alert the teaching faculty if this volume is greater than 8 μL.
- Add the appropriate volume of DEPC-treated water to a PCR tube.
- The total volume of RNA + DEPC-treated water should equal 8 μL.
- Add 1 μL of the oligo(dT)20 primer and 1 μL of the dNTP mix from the stock located at the front laboratory bench.
- Add the volume of your RNA solution that you calculated in Step #1.
- Incubate the RNA-primer mixture in the 65 °C water bath on the front laboratory bench for 5 min.
- Retrieve your tube and incubate on ice for 1 min.
- Prepare enough 'cDNA Synthesis Master Mix' for 2.1 reactions given the volumes of each component required for one reaction that are listed below. You will pipet the stock reagents at the front laboratory bench using filtered pipet tips.
- 2 μL of 10X RT buffer
- 4 μL of 25 mM MgCl2
- 2 μL of 0.1 M DTT
- 1 μL of RNaseOUT
- Add 9 μL of cDNA Synthesis Master Mix to each of you RNA-primer tubes and 1 μL of the SuperScriptIII RT from the tube located at the front laboratory bench.
- Run the 'RT' program on the thermocycler:
- Reactions will be incubated at 50 °C for 50 min, then terminated at 85 °C for 5 min.
- Retrieve your tubes, leave on ice for 1min and add 1 μL of RNase H. Incubate in the 37 °C incubator for 20 min.
- Your cDNA will be stored in the -20 °C freezer until the next laboratory session.
Reagents
- Qiashredder column (Qiagen)
- RNeasy mini kit (Qiagen)
- SuperScript III First-Strand kit, synthesis system for RT-PCR (Invitrogen)
- OligodT(20) 50uM
- 10mM dNTP mix
- 10X RT buffer
- 25mM MgCl2
- 0.1M DTT
- 200U/uL Superscript III RT
- 40U/uL RNaseOUT
- 2U/uL RNaseH
Next day: Journal club presentations
