Difference between revisions of "20.109(S16):Module 2"
MAXINE JONAS (Talk | contribs) (→Assignments) |
MAXINE JONAS (Talk | contribs) (→Overview) |
||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 12: | Line 12: | ||
===Overview=== | ===Overview=== | ||
| − | + | In the first module, your goal was to develop a tool that measures calcium concentration. For Module 2 you will use a research tool that was previously engineered to detect a molecular process and thereby address an interesting and important biological question. Specifically, you will investigate the importance of the DNA phosphokinase (DNA-PK) protein in a type of DNA repair called non-homologous end-joining, or NHEJ. Measuring DNA repair accurately and quantitatively may pave the way for certain cancer diagnostics and therapeutics. | |
| − | This time the DNA engineering has been done for you: the plasmid-based | + | This time the DNA engineering required to optimize the research tool for this specific project has been done for you: the plasmid-based reporter to measure NHEJ already exists, along with a novel version developed just for 20.109. In fact, you will be the first class to use the new version of the NHEJ reporter. With this reporter you will take a systems level view as you explore how different types of DNA damage are repaired by human fibroblasts harvested from a malignant glioblastoma. In your analysis you will interrogate the role of DNA-PK by completing your experiments with a 'wild-type' cell line and a mutant cell line lacking a subunit of DNA-PK, M059K and M059J, respectively. As you work through this module you will gain additional skills in analyzing and communicating information related to protein and cell-level assays. The culminating experiment will utilize flow cytometry, an amazing and infinitely useful technique that measures the fluorescence of individual cells. To evaluate class-wide trends in the flow data, you will learn and use basic statistical tools. |
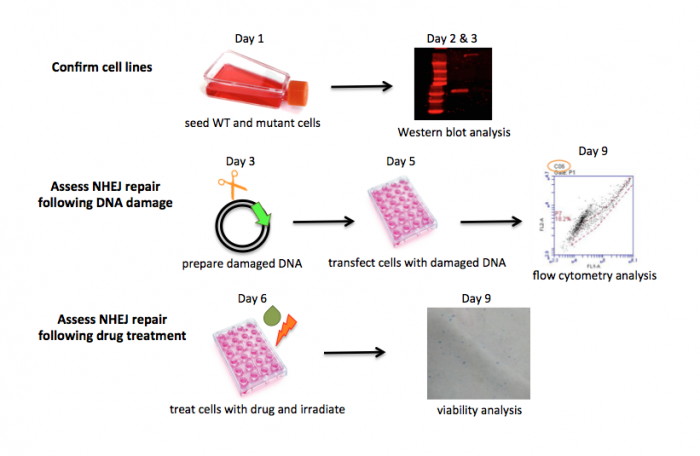
| − | + | The experiments you will complete in this module address three research questions. First, is DNA-PK present in the wild-type M059K cell line and, perhaps more importantly, is DNA-PK absent from the M059J mutant cell line. Second, how does the type of DNA damage impact NHEJ repair? Third, are there commercially available drugs that inhibit NHEJ? We will use a series of experiments to answer these questions as outlined in the overview schematic below. As you can see, we will be busy during Module 2... please keep notes and ask questions as we progress through the experiments to ensure you are prepared to write your research article. The toolkit you develop during the first two modules should leave you well poised to tackle the third and final module. | |
| − | [[Image: | + | [[Image:Sp16 M2 overview.png|thumb|700px|center|'''Overview of M2 experiments.''']] |
| + | |||
| + | We thank Agi Stachowiak, a former 20.109 technical instructor, as well as Alex Chaim and Zac Nagel from the Samson laboratory for their assistance in the development of this module, along with Marcus Parish for assistance with the irradiation assays. | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
Latest revision as of 16:45, 5 February 2016
Module 2
Lecturer: Leona Samson
Instructors: Noreen Lyell, Leslie McClain and Maxine Jonas
TA: Jing Zhang
Lab manager: Hsinhwa Lee
Overview
In the first module, your goal was to develop a tool that measures calcium concentration. For Module 2 you will use a research tool that was previously engineered to detect a molecular process and thereby address an interesting and important biological question. Specifically, you will investigate the importance of the DNA phosphokinase (DNA-PK) protein in a type of DNA repair called non-homologous end-joining, or NHEJ. Measuring DNA repair accurately and quantitatively may pave the way for certain cancer diagnostics and therapeutics.
This time the DNA engineering required to optimize the research tool for this specific project has been done for you: the plasmid-based reporter to measure NHEJ already exists, along with a novel version developed just for 20.109. In fact, you will be the first class to use the new version of the NHEJ reporter. With this reporter you will take a systems level view as you explore how different types of DNA damage are repaired by human fibroblasts harvested from a malignant glioblastoma. In your analysis you will interrogate the role of DNA-PK by completing your experiments with a 'wild-type' cell line and a mutant cell line lacking a subunit of DNA-PK, M059K and M059J, respectively. As you work through this module you will gain additional skills in analyzing and communicating information related to protein and cell-level assays. The culminating experiment will utilize flow cytometry, an amazing and infinitely useful technique that measures the fluorescence of individual cells. To evaluate class-wide trends in the flow data, you will learn and use basic statistical tools.
The experiments you will complete in this module address three research questions. First, is DNA-PK present in the wild-type M059K cell line and, perhaps more importantly, is DNA-PK absent from the M059J mutant cell line. Second, how does the type of DNA damage impact NHEJ repair? Third, are there commercially available drugs that inhibit NHEJ? We will use a series of experiments to answer these questions as outlined in the overview schematic below. As you can see, we will be busy during Module 2... please keep notes and ask questions as we progress through the experiments to ensure you are prepared to write your research article. The toolkit you develop during the first two modules should leave you well poised to tackle the third and final module.
We thank Agi Stachowiak, a former 20.109 technical instructor, as well as Alex Chaim and Zac Nagel from the Samson laboratory for their assistance in the development of this module, along with Marcus Parish for assistance with the irradiation assays.
Lab Links
M2D1: Introduction to cell strains and plating
M2D2: Begin Western protein analysis and choose system conditions
M2D3: Complete Western and prepare damaged DNA
M2D4: Journal Club I
Note: spring break occurs between Days 4 and 5.
M2D5: Cell preparation for DNA repair assays
M2D6: DNA repair assays
M2D7: Flow cytometry and paper discussion
M2D8: Journal Club II
M2D9: Data analysis