Difference between revisions of "20.109(F19):Module 1"
Noreen Lyell (Talk | contribs) (→Lab links: day by day) |
Noreen Lyell (Talk | contribs) (→Overview) |
||
| (7 intermediate revisions by one user not shown) | |||
| Line 11: | Line 11: | ||
==Overview== | ==Overview== | ||
| + | |||
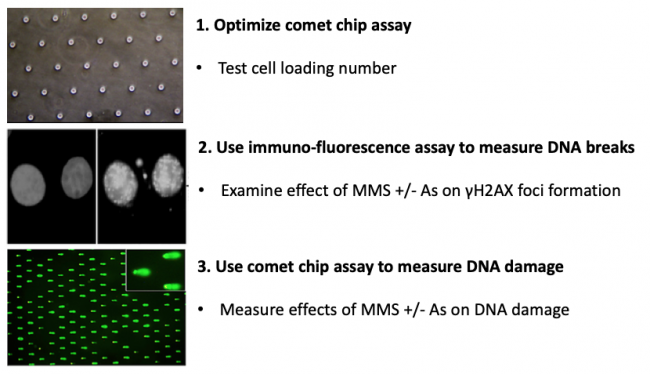
| + | Cancer is a disease of the genome. Cancer is caused by accumulated mutations in genes that lead to phenotypic advantages in the progression from normal to metastatic cancer. While we know a lot about the kinds of changes that cancer cells undergo, we do not yet fully understand where mutations come from in the first place. What is clear is that changes to the DNA structure (e.g., DNA damage) can cause carcinogenic mutations. Given that DNA damage causes mutations that drive cancer progression, it is important to have effective tools for measuring DNA damage. In this module, you will learn about two different approaches for quantifying DNA damage. The first method relies on physical changes to the DNA structure that impact its ability to migrate when electrophoresed. This is a direct measure of DNA damage. The second method relies on antibody recognition of changes to proteins that occur as a result of cell signaling that is triggered by DNA damage. This is an indirect measure of DNA damage. Given the potentially deadly consequences of DNA damage, it is a good thing that our cells have robust ways to repair their DNA. Being able to measure DNA damage means that we can study DNA repair, which turns out to be a very important variable when it comes to why some people get cancer, and others do not. | ||
| + | |||
| + | In addition to learning many fundamental biological and engineering concepts, you will of course learn many laboratory techniques. In particular, you will learn how to perform immunofluorescence, methods for quantitative image analysis, basic statistics, molecular pathway analysis, and much more. | ||
| + | |||
| + | An important focus of this module is public health. Of particular interest is the role that DNA damage plays in cancer susceptibility. There are several Superfund Sites in Massachusetts (these are sites that have high levels of legacy contaminants caused by prior industrial activities). In Wilmington, the Olin Superfund Site is contaminated with a DNA damaging agent (specifically a methylating agent called N-nitrosodimethylamine, or NDMA) as well as with arsenic. While there are published reports that arsenic might affect DNA repair, the possibility that these two exposures might act synergistically has not been fully explored. Here, you will play the role of a public health researcher with the goal of revealing possible combinatorial effects of a methylating agent and a metal. Results from these studies will help guide decision making with regard to environmental remediation. | ||
| + | |||
| + | In terms of specific experiments, in this module you will measure genomic instability using two techniques: a repair foci experiment (γH2AX immunofluorescence) and a high-throughput genome damage assay (CometChip). You will use these methods to assess the effect of exposure to contaminants known to cause DNA damage. Specifically, you will assess the levels of DNA damage caused by low concentrations of methyl methanosulfate (MMS) and arsenite (As) alone and in combination. MMS is a DNA methylating agent that causes DNA methylation lesions (MMS is a model agent that creates the same types of DNA damage as NDMA). Methylated bases are repaired by the base excision repair (BER) pathway. In the BER pathway, the damage is repaired by removing the damaged base, cleaving the backbone, replicating across the gap and ligating the remaining nick. There is evidence that arsenic inhibits key steps in the BER pathway. ''The prediction is therefore that As will lead to higher levels of MMS-induced DNA damage, raising the possibility that combined exposure an important risk to public health. For this module, you will use cutting edge genome damage assays to see for yourself if this is the case!'' | ||
| + | |||
| + | |||
| + | [[Image:Fa19 M1 overview.png|thumb|center|650px]] | ||
| + | |||
| + | <br style="clear:both" /> | ||
==Lab links: day by day== | ==Lab links: day by day== | ||
| − | M1D1: [[20.109(F19):Practice cell culture and begin sub-nuclear foci assay (Day1)| | + | M1D1: [[20.109(F19):Practice cell culture and begin sub-nuclear foci assay (Day1)| Learn optimal methods for culturing mammalian cells and initiate repair foci analysis experiment]]<br> |
| − | M1D2: [[20.109(F19):Treat cells for gamma-H2AX (Day2)| | + | M1D2: [[20.109(F19):Treat cells for gamma-H2AX (Day2)| Perform cell exposures for repair foci experiment and perform high-throughput genome damage pilot]]<br> |
| − | M1D3: [[20.109(F19):Complete gamma-H2AX assay and prepare CometChip (Day3)| | + | M1D3: [[20.109(F19):Complete gamma-H2AX assay and prepare CometChip (Day3)| Perform immunofluorescence assay for repair foci and optimize high-throughput genome damage assay]]<br> |
| − | M1D4: [[20.109(F19):Load cells into CometChip and apply treatments (Day4)| | + | M1D4: [[20.109(F19):Load cells into CometChip and apply treatments (Day4)| Perform immunofluorescence imaging for repair foci and expose cell for high-throughput genome damage assay]]<br> |
| − | M1D5: [[20.109(F19):Complete CometChip experiment (Day5)| | + | M1D5: [[20.109(F19):Complete CometChip experiment (Day5)| Quantify repair foci via network algorithm and complete high-throughput genome damage assay]]<br> |
| − | M1D6: [[20.109(F19):Image CometChip (Day6)| | + | M1D6: [[20.109(F19):Image CometChip (Day6)| Perform quantitative image analysis for the high-throughput genome damage assay]]<br> |
| − | M1D7: [[20.109(F19):Practice statistical analysis methods and complete data analysis (Day7)| | + | M1D7: [[20.109(F19):Practice statistical analysis methods and complete data analysis (Day7)| Complete data analysis and apply statistical methods]]<br> |
==Assignments== | ==Assignments== | ||
Latest revision as of 22:27, 10 September 2019
Contents
Module 1
Lecturer: Bevin Engelward
Instructors: Noreen Lyell, Leslie McClain and Becky Meyer
TAs: Colin Kim and Shelbi Parker
Lab manager: Hsinhwa Lee
Overview
Cancer is a disease of the genome. Cancer is caused by accumulated mutations in genes that lead to phenotypic advantages in the progression from normal to metastatic cancer. While we know a lot about the kinds of changes that cancer cells undergo, we do not yet fully understand where mutations come from in the first place. What is clear is that changes to the DNA structure (e.g., DNA damage) can cause carcinogenic mutations. Given that DNA damage causes mutations that drive cancer progression, it is important to have effective tools for measuring DNA damage. In this module, you will learn about two different approaches for quantifying DNA damage. The first method relies on physical changes to the DNA structure that impact its ability to migrate when electrophoresed. This is a direct measure of DNA damage. The second method relies on antibody recognition of changes to proteins that occur as a result of cell signaling that is triggered by DNA damage. This is an indirect measure of DNA damage. Given the potentially deadly consequences of DNA damage, it is a good thing that our cells have robust ways to repair their DNA. Being able to measure DNA damage means that we can study DNA repair, which turns out to be a very important variable when it comes to why some people get cancer, and others do not.
In addition to learning many fundamental biological and engineering concepts, you will of course learn many laboratory techniques. In particular, you will learn how to perform immunofluorescence, methods for quantitative image analysis, basic statistics, molecular pathway analysis, and much more.
An important focus of this module is public health. Of particular interest is the role that DNA damage plays in cancer susceptibility. There are several Superfund Sites in Massachusetts (these are sites that have high levels of legacy contaminants caused by prior industrial activities). In Wilmington, the Olin Superfund Site is contaminated with a DNA damaging agent (specifically a methylating agent called N-nitrosodimethylamine, or NDMA) as well as with arsenic. While there are published reports that arsenic might affect DNA repair, the possibility that these two exposures might act synergistically has not been fully explored. Here, you will play the role of a public health researcher with the goal of revealing possible combinatorial effects of a methylating agent and a metal. Results from these studies will help guide decision making with regard to environmental remediation.
In terms of specific experiments, in this module you will measure genomic instability using two techniques: a repair foci experiment (γH2AX immunofluorescence) and a high-throughput genome damage assay (CometChip). You will use these methods to assess the effect of exposure to contaminants known to cause DNA damage. Specifically, you will assess the levels of DNA damage caused by low concentrations of methyl methanosulfate (MMS) and arsenite (As) alone and in combination. MMS is a DNA methylating agent that causes DNA methylation lesions (MMS is a model agent that creates the same types of DNA damage as NDMA). Methylated bases are repaired by the base excision repair (BER) pathway. In the BER pathway, the damage is repaired by removing the damaged base, cleaving the backbone, replicating across the gap and ligating the remaining nick. There is evidence that arsenic inhibits key steps in the BER pathway. The prediction is therefore that As will lead to higher levels of MMS-induced DNA damage, raising the possibility that combined exposure an important risk to public health. For this module, you will use cutting edge genome damage assays to see for yourself if this is the case!
Lab links: day by day
M1D1: Learn optimal methods for culturing mammalian cells and initiate repair foci analysis experiment
M1D2: Perform cell exposures for repair foci experiment and perform high-throughput genome damage pilot
M1D3: Perform immunofluorescence assay for repair foci and optimize high-throughput genome damage assay
M1D4: Perform immunofluorescence imaging for repair foci and expose cell for high-throughput genome damage assay
M1D5: Quantify repair foci via network algorithm and complete high-throughput genome damage assay
M1D6: Perform quantitative image analysis for the high-throughput genome damage assay
M1D7: Complete data analysis and apply statistical methods
Assignments
Data summary
Mini-presentation
References
CometChip: A high-throughput 96-well platform for measuring DNA damage in microarrayed human cells. Journal of Visualized Experiments. (2014) 92: 1-11.
- A video of the procedure is linked here.
CometChip: Single-cell microarray for high-throughput detection of DNA damage. Methods in Cell Biology. (2012) 112: 247-268.