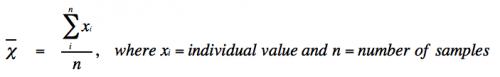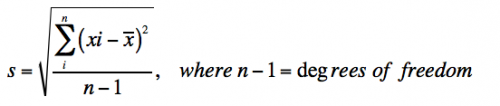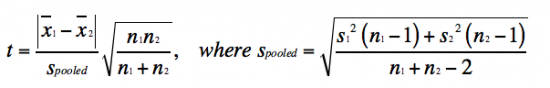20.109(F19):Practice statistical analysis methods and complete data analysis (Day7)
Contents
Introduction
Today is the final laboratory session for Module 1! You have completed all of the bench work for your research; however, there is still data analysis to complete for your experiments. In addition to plotting the data, you will complete statistical analysis to determine the significance of your results.
Statistics are mathematical tools used to analyze, interpret, and organize data. The specific tools that you will use are confidence intervals (CI) and the Student's t-test. To begin, review the following definitions:
- Mean (or average) is defined as:
- With infinite data, the mean (χι) approaches the true mean (μ).
- Standard deviation measures the variation in the data and is defined as:
- With infinite data, the standard deviation (s) approaches the true standard deviation (σ).
Because standard deviation is only justified when sufficient data have been collected to generate a normal curve, you will use confidence intervals to report the likelihood that your results predict the true mean. A confidence interval is a defined interval that is calculated to define the true mean to a specified level of confidence. Simply, it is possible to define a range in your data set that likely contains the true mean based on the calculated mean.
- Confidence interval is defined as:
In your data, you should use the CI to generate error bars due the low n. Be sure to report which confidence level was used to calculate the intervals reported. So, what does this all mean in regard to the data you will report? As an example, if the calculated χι of a data set equals 80 au there is a 95% chance the μ is between 50 au and 110 au, where au = arbitrary units. And how does this relate to s? If you know the μ, the σ represents a 68% confidence interval.
Lastly, you will use Student's t test to report if your data are statistically different between treatments.
- Student's t test is defined as:
The value you calculate with the Student's t test equation is referred to as tcalculated. This tcalculated value is compared to the ttabulated value in the the t table, according to the appropriate n - 1 using the p-value for the two-tailed distribution (which assumes that you do not know how the data will shift). If the tcalculated value is greater than the ttabulated, then the data sets are significantly different at the specific p-value. So, what does this all mean in regard to the data you will report? As an example, if the tcalculated for a data set with n - 1 = 10 is 3 (given that the ttabulated is 2.228), then the data sets are different with a p-value ≤ 0.05. Which means that there is less that a 5% chance that the data sets are the same.
Protocols
Part 1: Practice statistical analysis
Review these data from an experiment where cells were exposed to increasing amounts of radiation. Your goal is to determine if a statistically significant amount of DNA damage was induced. For the purpose of this exercise, the values in the spreadsheet are in arbitrary units of 'DNA damage', where the higher numbers indicate more damage.
When interpreting the statistics, consider how you may use the information to convince someone that the DNA damage was significant. You may find this spreadsheet, originally created by Prof. Bevin Engelward and modified by the 20.109 staff, helpful for this exercise. At a minimum, you should post a bar plot of the data with 95% confidence intervals and indicate if there is a statistically significant difference (i.e. provide a p-value) between conditions in your Benchling notebook.
Part 2: Complete data analysis
Use the tools above to analyze the data for your CometChip and gamma-H2AX experiments. The figures / analyses in your Data summary should include measures of variability (i.e. confidence intervals) and significance (i.e. p-values).
Part 3: Draft first data slide for Data summary
To get a headstart and feedback from the teaching faculty, you will draft the first data slide for your Summary today in class. With your partner, use the template below to generate a slide that presents the cell loading data used to generate your figure in the previous session.
Individually you each created figures (with a title and caption) using the cell loading data. In this exercise you will come together with your laboratory partner to decide how to best present these data and then include the fluorescence microscopy information. Because of the added information, you will need to update / modify your caption and possibly the title! In addition, complete the results / discussion bullets using the questions in the template as a guide.
Part 4: Overlay H2AX channels to make a single color image
Now you will use the folder called "Three channel images" to put together some representative images for your report. For these image stacks, images were taken in three different fluorescence channels: DAPI, FITC, and TxRed. The DAPI channel contains images of the nuclei from the DAPI stain, the FITC channel contains images of the EdU stain, and the TxRed channel contains images of the gamma H2AX immunofluorescence.
- Open one image file for one condition in ImageJ. Choose the 'Image' dropdown menu, select Stacks, then choose Stack to Images. This splits your one image stack into 3 separate files.
- Select Image -> Color -> Merge channels. A pop-up window will appear.
- Assign colors to your tif images using the dropdown selection of file names. Choose *none* for all colors that are not represented in your image. Traditionally DAPI is represented as blue, AlexaFluor488 is represented as green, and AlexaFluor594 is represented as red.
- Make sure that the bottom 3 check boxes are not selected and click OK.
- The three files you assigned colors are now overlayed into one color RGB file. You must save and name this file before closing the window to avoid losing the color overlay.
- Don't forget to include a scale bar on your image. The images were taken at 60x magnification, with each pixel representing 0.108um.
- Select Analyze -> Set Scale : "Distance in pixels" is 1, "Known distance" is 0.108, "Pixel aspect ratio" is 1, "Unit of length" is um, and select OK
- Select Analyze -> Tools -> Scale bar: Choose appropriate settings (e.g. 50 um in width, and you can eliminate text here and include the scale related text in your caption).
Next day: Complete in silico cloning of pdCas9