Difference between revisions of "20.109(F19):Image CometChip (Day6)"
Noreen Lyell (Talk | contribs) (→Part 1: Communication Lab workshop) |
Noreen Lyell (Talk | contribs) (→Part 3: Image CometChips) |
||
| Line 19: | Line 19: | ||
You can submit the updated version of the assignment by 10 pm tonight for grading. | You can submit the updated version of the assignment by 10 pm tonight for grading. | ||
| − | ===Part 3: | + | ===Part 3: Process microscopy images for DNA damage experiment=== |
| − | For timing reasons, | + | For timing reasons, the teaching faculty imaged your CometChip for the DNA damage experiment. To ensure you understand the process you will see a demonstration of the process during laboratory today. |
| + | |||
| + | To analyze your DNA damage experiment results, download your CometChip images from the [[ Class data tab]] or obtain images from an instructor. Create (or copy and paste) the folder of your team images into the Documents\MATLAB\CometChip Analysis directory of the lab computer. <br> | ||
| + | |||
| + | You will complete the following processes twice (stack images, optimize analysis parameters, measure tail lengths, export to Excel)--once for each CometChip. You should end up with one Excel file for each chip. | ||
| + | |||
| + | '''Stack images using ImageJ''' | ||
| + | #The script you will run will combine images from the same well into a stacked image and rename the files for the subsequent Matlab script to recognize them. | ||
| + | #Open ImageJ from Applications. | ||
| + | #Go to Plugins → Macros → Run... | ||
| + | #Select script "GenImageStacks_singleimage.txt" within the Documents\MATLAB\CometChip Analysis directory | ||
| + | #Choose the appropriate source directory that contains your image files. | ||
| + | #*Find in Documents\MATLAB\CometChip Analysis directory | ||
| + | #Create a destination directory by selecting New Folder and naming the folder appropriately (eg. "180927M059_H2O2_stacked"). | ||
| + | #Click Choose. | ||
| + | #*ImageJ will create the stacked images in ~2 min. | ||
| + | #*Please do not hit any additional keys until this process is completed. | ||
| + | #Confirm that the stacks are in the destination directory. | ||
| + | #*Open the directory you just created, containing your stacked images. | ||
| + | #*You should see one .tif image stack per well. | ||
| + | #Close ImageJ. | ||
| + | |||
| + | '''Optimize analysis parameters using MATLAB''' | ||
| + | #Open MATLAB from Applications. | ||
| + | #*Be sure that "CometChip Analysis" is the current folder (double click on the "CometChip Analysis" folder from within the "Current Folder" window). | ||
| + | #In the command window, type in "guicometanalyzer" press enter to run the script. | ||
| + | #In the MultiWell Comet Analyzer window: | ||
| + | #*Check that the Pixel Calibration value is 1.61. | ||
| + | #*Check that the Image Rotation value is 0 if your tails are to the right. Image Rotation should be 180 if your tails are going to the left. | ||
| + | #*Check that the Array Spacing (μm) is 240. | ||
| + | #*Check that the Head Diameter value is 20. | ||
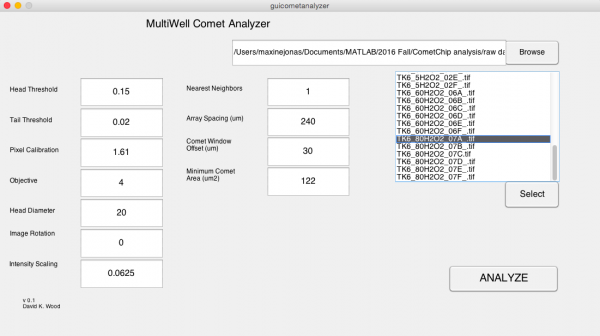
| + | #*Confirm the settings with the image below.[[Image:Fa16 M1D5 comet analyzer window.png|thumb|center|600px|'''Multiwell Comet Analyzer window.''']] | ||
| + | #Click Browse. | ||
| + | #*Select the stacked image directory you created in the previous section. | ||
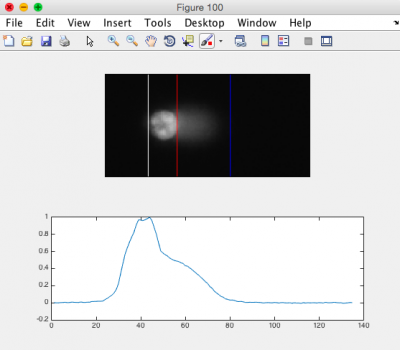
| + | #*Click Open.[[Image:Fa16 M1D5 comet head tail parameters.png|thumb|right|400px|'''CometChip analysis showing differentiation between head and tail.''']] | ||
| + | #From the image stack files loaded to the MultiWell Comet Analyzer window, choose '''ONE image''' that is expected to show DNA damage. You are just using this image to optimize the analysis parameters. | ||
| + | #*The chosen file should be highlighted. | ||
| + | #*Click Select. | ||
| + | #Click Analyze. | ||
| + | #Click "Yes" when the dialog box appears and asks if you want to run the program in "debug mode." | ||
| + | #*Please do not hit any additional keys until this process is completed. | ||
| + | #*In the MATLAB command window you will see the number of comets to be analyzed. The same number of figures should be generated. | ||
| + | #Review the generated images. | ||
| + | #*Ensure that the head of the comet is bracketed by a white line on the left, and a red line on the right. The tail should be bracketed by the red line and a blue line on the right. See the image to the right for an example. | ||
| + | #*You may need to adjust the "Tail Threshold" value so that the tail is identified appropriately and/or the "Head Threshold" value so that the head is identified appropriately. | ||
| + | #*Note: it is okay if some of the images do not appear correct according to the above criteria; however, the majority of your images should be correct. | ||
| + | #In the Command Window type "close all." | ||
| + | #If you need to adjust your parameters, repeat this process again until you find the optimal parameters for your analysis. | ||
| + | #*Find the guicometanalzyer window. | ||
| + | #*Adjust parameters as needed and repeat steps above by continuing to run in "debug mode." | ||
| + | #*Type "close all" when done, but leave Matlab open. | ||
| + | |||
| + | '''Measure head and tail lengths using MATLAB''' | ||
| + | #Find the guicometanalyzer window | ||
| + | #*If it has disappeared, run the script again by typing "guicometanalyzer" in the command window. | ||
| + | #In the MultiWell Comet Analyzer window, confirm the parameters as above. | ||
| + | #*Click Browse. | ||
| + | #*Select the stacked image directory you created with ImageJ. | ||
| + | #*Click Open. | ||
| + | #Highlight '''all stack files''' in the MultiWell Comet Analyzer window. | ||
| + | #*Click Select. | ||
| + | #Click Analyze. | ||
| + | #Click '''"No"''' when the dialog box appears and asks if you want to run the program in "debug mode." | ||
| + | #Confirm that there is a .txt file for each stack in your folder. | ||
| + | #*Be sure to keep MATLAB open. | ||
| + | |||
| + | '''Compile results and export to Excel''' | ||
| + | #In the command window, type "comettoexcel" and press enter to run the script. | ||
| + | #When prompted, select the directory containing all the txt files that were output by guicometanalyzer and click "Open." | ||
| + | #The script will compile all your data, calculate the median values for each well, and export the data in an Excel file. | ||
| + | #Next, a dialog box will prompt you to save the Excel file. | ||
| + | #Please choose an appropriate directory and file name to save your data. | ||
| + | #Click "Save" and ensure your data has saved correctly. | ||
| + | |||
| + | '''Post your final Excel spreadsheets on the [[20.109(F19):Class data| Class data tab]]. | ||
===Part 4: Group paper discussion=== | ===Part 4: Group paper discussion=== | ||
Revision as of 19:38, 22 August 2019
Contents
Introduction
The goal for today is to get you started on the Data summary!
Protocols
Part 1: Communication Lab workshop
Our communication instructors, Dr. Sean Clarke and Dr. Prerna Bhargava, will join us today for a discussion on designing effective figures and captions.
Part 2: Modify homework
Use the information provided during the Communication Lab workshop to edit your Figure / Caption homework assignment. Also, the teaching faculty will meet with each team to provide specific feedback and comments.
You can submit the updated version of the assignment by 10 pm tonight for grading.
Part 3: Process microscopy images for DNA damage experiment
For timing reasons, the teaching faculty imaged your CometChip for the DNA damage experiment. To ensure you understand the process you will see a demonstration of the process during laboratory today.
To analyze your DNA damage experiment results, download your CometChip images from the Class data tab or obtain images from an instructor. Create (or copy and paste) the folder of your team images into the Documents\MATLAB\CometChip Analysis directory of the lab computer.
You will complete the following processes twice (stack images, optimize analysis parameters, measure tail lengths, export to Excel)--once for each CometChip. You should end up with one Excel file for each chip.
Stack images using ImageJ
- The script you will run will combine images from the same well into a stacked image and rename the files for the subsequent Matlab script to recognize them.
- Open ImageJ from Applications.
- Go to Plugins → Macros → Run...
- Select script "GenImageStacks_singleimage.txt" within the Documents\MATLAB\CometChip Analysis directory
- Choose the appropriate source directory that contains your image files.
- Find in Documents\MATLAB\CometChip Analysis directory
- Create a destination directory by selecting New Folder and naming the folder appropriately (eg. "180927M059_H2O2_stacked").
- Click Choose.
- ImageJ will create the stacked images in ~2 min.
- Please do not hit any additional keys until this process is completed.
- Confirm that the stacks are in the destination directory.
- Open the directory you just created, containing your stacked images.
- You should see one .tif image stack per well.
- Close ImageJ.
Optimize analysis parameters using MATLAB
- Open MATLAB from Applications.
- Be sure that "CometChip Analysis" is the current folder (double click on the "CometChip Analysis" folder from within the "Current Folder" window).
- In the command window, type in "guicometanalyzer" press enter to run the script.
- In the MultiWell Comet Analyzer window:
- Check that the Pixel Calibration value is 1.61.
- Check that the Image Rotation value is 0 if your tails are to the right. Image Rotation should be 180 if your tails are going to the left.
- Check that the Array Spacing (μm) is 240.
- Check that the Head Diameter value is 20.
- Confirm the settings with the image below.
- Click Browse.
- Select the stacked image directory you created in the previous section.
- Click Open.
- From the image stack files loaded to the MultiWell Comet Analyzer window, choose ONE image that is expected to show DNA damage. You are just using this image to optimize the analysis parameters.
- The chosen file should be highlighted.
- Click Select.
- Click Analyze.
- Click "Yes" when the dialog box appears and asks if you want to run the program in "debug mode."
- Please do not hit any additional keys until this process is completed.
- In the MATLAB command window you will see the number of comets to be analyzed. The same number of figures should be generated.
- Review the generated images.
- Ensure that the head of the comet is bracketed by a white line on the left, and a red line on the right. The tail should be bracketed by the red line and a blue line on the right. See the image to the right for an example.
- You may need to adjust the "Tail Threshold" value so that the tail is identified appropriately and/or the "Head Threshold" value so that the head is identified appropriately.
- Note: it is okay if some of the images do not appear correct according to the above criteria; however, the majority of your images should be correct.
- In the Command Window type "close all."
- If you need to adjust your parameters, repeat this process again until you find the optimal parameters for your analysis.
- Find the guicometanalzyer window.
- Adjust parameters as needed and repeat steps above by continuing to run in "debug mode."
- Type "close all" when done, but leave Matlab open.
Measure head and tail lengths using MATLAB
- Find the guicometanalyzer window
- If it has disappeared, run the script again by typing "guicometanalyzer" in the command window.
- In the MultiWell Comet Analyzer window, confirm the parameters as above.
- Click Browse.
- Select the stacked image directory you created with ImageJ.
- Click Open.
- Highlight all stack files in the MultiWell Comet Analyzer window.
- Click Select.
- Click Analyze.
- Click "No" when the dialog box appears and asks if you want to run the program in "debug mode."
- Confirm that there is a .txt file for each stack in your folder.
- Be sure to keep MATLAB open.
Compile results and export to Excel
- In the command window, type "comettoexcel" and press enter to run the script.
- When prompted, select the directory containing all the txt files that were output by guicometanalyzer and click "Open."
- The script will compile all your data, calculate the median values for each well, and export the data in an Excel file.
- Next, a dialog box will prompt you to save the Excel file.
- Please choose an appropriate directory and file name to save your data.
- Click "Save" and ensure your data has saved correctly.
Post your final Excel spreadsheets on the Class data tab.
Part 4: Group paper discussion
To better understand the structure and content for written communications in science, we will discuss the following research article:
Weingeist, D. M., et al. Single-cell microarray enables high-throughput evaulation of DNA doubl-strand breaks and DNA repair inhibitors. (2013) Cell Cycle. 126:907-915.
Our paper discussion will be guided by all that you have learned about how to write a cohesive story that clearly reports the data and provides strong support for the conclusions made about the data. During the paper discussion, everyone is expected to participate - either by volunteering or by being called upon!
Introduction
Remember the key components of an introduction:
- What is the big picture?
- Is the importance of this research clear?
- Are you provided with the information you need to understand the research?
- Do the authors include a preview of the key results?
Results
Carefully examine the figures. First, read the captions and use the information to 'interpret' the data presented within the image. Second, read the text within the results section that describes the figure.
- Do you agree with the conclusion(s) reached by the authors?
- What controls were included and are they appropriate for the experiment performed?
- Are you convinced that the data are accurate and/or representative?
Discussion
Consider the following components of a discussion:
- Are the results summarized?
- Did the authors 'tie' the data together into a cohesive and well-interpreted story?
- Do the authors overreach when interpreting the data?
- Are the data linked back to the big picture from the introduction?
Next day: Practice statistical analysis methods and complete data analysis