20.109(F19):Begin Western blot analysis (Day3)
Contents
Introduction
Electrophoresis is a technique that separates large molecules by size using an applied electrical field and a sieving matrix. DNA, RNA and proteins are the molecules most often studied with this technique; agarose and acrylamide gels are the two most common sieves. The molecules to be separated enter the matrix through a well at one end and are pulled through the matrix when a current is applied across it. The larger molecules get entwined in the matrix and are stalled; the smaller molecules wind through the matrix more easily and travel farther away from the well. The distance a nucleic acid or amino acid fragment travels is inversely proportional to the log of its length. Over time fragments of similar length accumulate into “bands” in the gel.
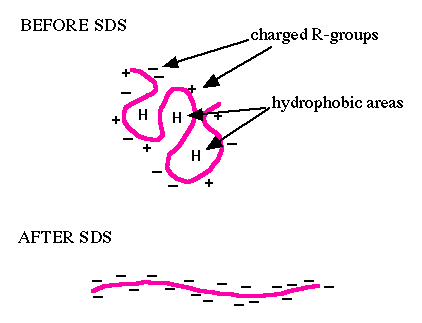
You will use sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to evaluate FKPB12. SDS is an ionic surfactant (or detergent), which denatures the proteins and coats them with a negative charge. Since denatured proteins are linear, they will move through the gel at a speed inversely proportional to their molecular weight, just like DNA on agarose gels. (Non-denatured proteins run according to their molecular weight, shape, and charge.) You will run a reference ladder containing proteins of known molecular weight and amount to determine the size and concentration of your purified FKBP12 protein. When running uninduced and induced samples side-by-side, you should see a protein band at the expected molecular weight for FKBP12 (~12 kDa). Following electrophoresis, you will transfer the cellular proteins within the lysate from the polyacrylamide gel to a membrane. Because membranes are more stabile and easier to handle, this is used to 'blot' for proteins of interest in the mix of cellular proteins. In a Western blot, an antibody is used to tag the protein of interest providing information on whether or not the protein is present in a sample, and if present, at what abundance.For Western blot analysis, a high quality antibody can have a relatively low affinity for its target protein. This is because the target is localized and concentrated on a blot, allowing the antibody to bind using both antibody “arms” thereby strengthening the association. Even an antibody that is loosely bound to the blot under these circumstances may dissociate then re-associate quickly since the local concentration of the target protein is high. The lower limit for protein detection is approximately 1 ng/lane, a value that varies with the size of the protein to be detected and the Western blotting apparatus that is used. For most polyacrylamide gels, the protein capacity for each lane is 100 to 200 μg (that would be 20 μL of a 5-10 μg/μL protein preparation). Thus, 1 ng represents a protein that is approximately 0.0005-0.001% of the total cellular protein (1 ng out of 100,000-200,000 ng). Proteins that make up a more significant fraction of the total protein population will be easier to detect.
Protocols
In the previous laboratory session, you snap froze your samples which were then stored at -80 °C.
Part 1: Lyse cells
- Retrieve your samples from the front laboratory bench and place them in the 25 °C heatblock.
- Your samples will thaw in the block.
- When thawed, transfer to an ice bucket.
- Vortex your samples, then centrifuge the lysate at 20,000 g for 20 min in the 4 °C cold room.
- Alert the teaching faculty when you are ready to centrifuge.
- Following centrifugation, leave your samples on ice.
- Be careful not to disturb the pellet!
- Transfer 90 μL of the lysate into a fresh 1.5 mL microcentrifuge tube.
Part 2: Electrophorese cellular proteins
- Add 15 μL of Laemmli sample buffer to each of your samples.
- Heat for 10 min at 70 °C using the heatblock on the front laboratory bench.
- If there is significant condensation at the lid of your tubes after heating, spin your samples before loading.
- You will load all samples and two molecular weight standards.
- A pre-stained ladder will be used to track the migration of your samples through the polyacrylamide gel.
- An unstained ladder with bands of known amounts of protein will be used to estimate protein concentration in your samples.
- Record the order in which you will load your samples and molecular weight standards in the polyacrylamide gel.
- When you are ready to load your samples, alert the teaching faculty.
- Please watch the demonstration closely to ensure your samples are correctly loaded and the polyacrylamide gel is not damaged during loading.
- Your samples will be electrophoresed at 200 V for 30-45 min.
- Following electrophoresis, use the spatula to carefully pry apart the plates that encase your polyacrylamide gel.
- Using wet gloves, transfer your polyacrylamide gel to a staining box and add enough dH2O to cover the gel.
- Wash the gel for 5 min at room temperature on the rotating table.
- Empty the water from the staining box in the sink.
- Be careful that the gel does not fall into the sink!
- Repeat Steps #12-13 a total of 3 times.
Part 3: Transfer proteins to membrane
The teaching faculty will take two teams at a time to the iBlot in Prof. Griffith's laboratory for the transfer step. Please be patient until it is your turn.
Reagents list
- SDS-PAGE gels, 4-20% polyacrylamide gel in Tris-HCl (from BioRad)
- Tris, glycine, SDS (TGS) buffer: 25 mM Tris, 192 mM glycine, 0.1% (w/v) SDS, pH 8.3 (from BioRad)
- Kaleidoscope marker (from BioRad)
- band information can be found here
- unstained marker (from BioRad)
- band information can be found here
- iBlot 2 transfer stack (from ThermoFischer)
- nitrocellulose membrane
Next day: Complete antibody staining for Western blot analysis