Difference between revisions of "20.109(F16):Data analysis (Day7)"
Noreen Lyell (Talk | contribs) (→Introduction) |
MAXINE JONAS (Talk | contribs) (→Part 2: Image processing for H2AX assay) |
||
| (27 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
==Introduction== | ==Introduction== | ||
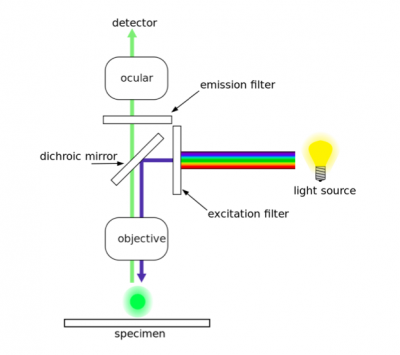
| + | As a brief reminder, two antibodies were used in the gamma-H2AX assay. The first antibody, or primary antibody, was anti-gamma-H2AX and raised in a mouse. The secondary antibody was anti-mouse and raised in a goat, more importantly, this molecule is conjugated to a fluorescent dye tag called Alexa Fluor 488. The Alexa Fluor 488 tag is a bright, green fluorescent dye that is excited at 488 nm. To visualize the abundance of double-strand breaks in your H2AX assay samples, we will use fluorescence microscopy. | ||
| + | |||
[[Image:Fa16 M1D7 fluorescence microscope.png|thumb|center|400px|'''Diagram of a fluorescence microscope.''']] | [[Image:Fa16 M1D7 fluorescence microscope.png|thumb|center|400px|'''Diagram of a fluorescence microscope.''']] | ||
| + | |||
| + | In fluorescence microscopy the specimen is illuminated with a wavelength of light specific to the excitation of the fluorescent tag used to target the feature of interest. The excitation wavelength is absorbed by the fluorescent tag, which causes it to emit light at a longer, less energetic wavelength. Typically, fluorescence microscopes used in biology are an epifluorescence type with a single light path (the objective) for excitation and emission detection, as depicted in the diagram above. | ||
| + | |||
| + | Fluorescence, or epifluorescence, microscopes are composed of a light source, an excitation filter, a dichroic mirror, and an emission filter. The filters and the dichroic mirror are specific to the spectral excitation and emission characteristics of the fluorescent tag. To visualize fluorescence, light at the excitation wavelength is focused on the sample. The light emission from the sample is focused by the objective to a detector. | ||
| + | |||
| + | Today you will image the samples you tested using the gamma-H2AX assay and complete a short summary that qualitatively interprets the data. | ||
==Protocols== | ==Protocols== | ||
===Part 1: Visualize H2AX assay results=== | ===Part 1: Visualize H2AX assay results=== | ||
| − | #Aspirate the secondary antibody solution. | + | #Make sure to have TBS solution available before you start. Aspirate the secondary antibody solution off the coverslip and immediately add 150 μL of TBS. Do not let the coverslips dry out during this process. |
| − | #To | + | #To complete the post secondary wash, add 150 μL of TBS per coverslip, let incubate at room temperature for 3 min covered, then aspirate. |
| − | #*Repeat this step | + | #*Repeat this step twice. |
| − | + | #Obtain glass slides from the front laboratory bench and label your slides with all of your experimental information and group name, add 5 μL of mounting media to the slide. | |
| − | #Obtain | + | #Aspirate the final TBS wash and using tweezers place the coverslip cell-side down on the mounting media "spot" on the microscope slide. Try your best to avoid bubbles by slowly placing the coverslip over the mounting media. |
| − | + | #*The cell-side of the coverslide is the side that was facing up in the staining chamber. | |
| − | + | #Complete Steps #3-4 for coverslips from the H<sub>2</sub>O<sub>2</sub> treated and control wells for both M059K and M059J cell lines. | |
| − | + | ||
| − | + | ||
| − | # | + | |
| − | #*The cell-side of the coverslide is the side that was facing up in the | + | |
| − | #Complete Steps # | + | |
#Alert the teaching faculty when all four microscope slides are ready and you will be escorted to the microscope in the Engelward laboratory. | #Alert the teaching faculty when all four microscope slides are ready and you will be escorted to the microscope in the Engelward laboratory. | ||
| − | ===Part 2: Data analysis for H2AX assay=== | + | ===Part 2: Image processing for H2AX assay=== |
| + | Once you have taken representative images of your two experimental condition slides and two control condition slides you will need to process the image files using ImageJ. | ||
| + | #Open both the DAPI and AlexaFluor488 image files for one condition in ImageJ. These images are currently a 16-bit tif file so they are both black and white. | ||
| + | #Choose the 'Image' dropdown menu, select color, then select merge channels. A pop-up window will appear. | ||
| + | #Assign colors to your 16-bit tif images using the dropdown selection of file names. Choose *none* for all colors that are not represented in your image. Traditionally DAPI is represented as blue and AlexaFluor488 is represented as green. | ||
| + | #Make sure that the bottom 3 check boxes are not selected and click OK. | ||
| + | #The two files you assigned colors are now overlayed into one color RGB file. You must save and name this file before closing the window to avoid losing the color overlay. | ||
| + | Repeat these steps for 'M059K untreated', 'M059K 1h H<sub>2</sub>O<sub>2</sub>', 'M059J untreated', and 'M059J 1h H<sub>2</sub>O<sub>2</sub>'. | ||
| + | |||
| + | ===Part 3: Data analysis for H2AX assay=== | ||
| + | The information pertaining to the H2AX assay '''will not''' be included in your [[20.109(F16): M1 Data Summary| M1 Data Summary]]. Instead, you will use these data to generate a single page write-up using the format required for the Data Summary. To help you practice crafting the 'data slides' for your Data Summary, you will prepare the data you collected in the H2AX assay as a figure (with title and caption!) and write corresponding results and interpretation bullets. | ||
| + | |||
| + | Use the example below as you prepare this assignment. Note: This will be counted in your homework grade and is due tonight at 10 pm (Thursday, October 6 or Friday, October 7, according to your laboratory section). You should follow the format guideline specified for the M1 Data Summary. Use your extra time in class and ask the teaching faculty any questions that may arise! | ||
| + | |||
| + | [[Image:Fa16 M1D7 data slide template format v2.png|thumb|center|650px|'''Template for data slide.''' Use this as a guide to complete the M1D7 in-class homework assignment and for the data slides that within the M1 Data Summary.]] | ||
| − | ==Reagents== | + | ==Reagents list== |
| + | *Tris buffer saline (TBS, Biorad) | ||
| + | *Mounting media ProLong gold with DAPI (Molecular Probes) | ||
==Navigation links== | ==Navigation links== | ||
| − | Next day: [[ | + | Next day: [[20.109(F16):Complete in silico cloning (Day1)| Module 2 begins!]] |
| − | Previous day: [[20.109(F16):Complete immuno-fluorescence assay (Day6)| | + | Previous day: [[20.109(F16):Complete immuno-fluorescence assay (Day6)| Query DNA repair capacity in tumor cells]] |
Latest revision as of 16:39, 6 October 2016
Contents
Introduction
As a brief reminder, two antibodies were used in the gamma-H2AX assay. The first antibody, or primary antibody, was anti-gamma-H2AX and raised in a mouse. The secondary antibody was anti-mouse and raised in a goat, more importantly, this molecule is conjugated to a fluorescent dye tag called Alexa Fluor 488. The Alexa Fluor 488 tag is a bright, green fluorescent dye that is excited at 488 nm. To visualize the abundance of double-strand breaks in your H2AX assay samples, we will use fluorescence microscopy.
In fluorescence microscopy the specimen is illuminated with a wavelength of light specific to the excitation of the fluorescent tag used to target the feature of interest. The excitation wavelength is absorbed by the fluorescent tag, which causes it to emit light at a longer, less energetic wavelength. Typically, fluorescence microscopes used in biology are an epifluorescence type with a single light path (the objective) for excitation and emission detection, as depicted in the diagram above.
Fluorescence, or epifluorescence, microscopes are composed of a light source, an excitation filter, a dichroic mirror, and an emission filter. The filters and the dichroic mirror are specific to the spectral excitation and emission characteristics of the fluorescent tag. To visualize fluorescence, light at the excitation wavelength is focused on the sample. The light emission from the sample is focused by the objective to a detector.
Today you will image the samples you tested using the gamma-H2AX assay and complete a short summary that qualitatively interprets the data.
Protocols
Part 1: Visualize H2AX assay results
- Make sure to have TBS solution available before you start. Aspirate the secondary antibody solution off the coverslip and immediately add 150 μL of TBS. Do not let the coverslips dry out during this process.
- To complete the post secondary wash, add 150 μL of TBS per coverslip, let incubate at room temperature for 3 min covered, then aspirate.
- Repeat this step twice.
- Obtain glass slides from the front laboratory bench and label your slides with all of your experimental information and group name, add 5 μL of mounting media to the slide.
- Aspirate the final TBS wash and using tweezers place the coverslip cell-side down on the mounting media "spot" on the microscope slide. Try your best to avoid bubbles by slowly placing the coverslip over the mounting media.
- The cell-side of the coverslide is the side that was facing up in the staining chamber.
- Complete Steps #3-4 for coverslips from the H2O2 treated and control wells for both M059K and M059J cell lines.
- Alert the teaching faculty when all four microscope slides are ready and you will be escorted to the microscope in the Engelward laboratory.
Part 2: Image processing for H2AX assay
Once you have taken representative images of your two experimental condition slides and two control condition slides you will need to process the image files using ImageJ.
- Open both the DAPI and AlexaFluor488 image files for one condition in ImageJ. These images are currently a 16-bit tif file so they are both black and white.
- Choose the 'Image' dropdown menu, select color, then select merge channels. A pop-up window will appear.
- Assign colors to your 16-bit tif images using the dropdown selection of file names. Choose *none* for all colors that are not represented in your image. Traditionally DAPI is represented as blue and AlexaFluor488 is represented as green.
- Make sure that the bottom 3 check boxes are not selected and click OK.
- The two files you assigned colors are now overlayed into one color RGB file. You must save and name this file before closing the window to avoid losing the color overlay.
Repeat these steps for 'M059K untreated', 'M059K 1h H2O2', 'M059J untreated', and 'M059J 1h H2O2'.
Part 3: Data analysis for H2AX assay
The information pertaining to the H2AX assay will not be included in your M1 Data Summary. Instead, you will use these data to generate a single page write-up using the format required for the Data Summary. To help you practice crafting the 'data slides' for your Data Summary, you will prepare the data you collected in the H2AX assay as a figure (with title and caption!) and write corresponding results and interpretation bullets.
Use the example below as you prepare this assignment. Note: This will be counted in your homework grade and is due tonight at 10 pm (Thursday, October 6 or Friday, October 7, according to your laboratory section). You should follow the format guideline specified for the M1 Data Summary. Use your extra time in class and ask the teaching faculty any questions that may arise!
Reagents list
- Tris buffer saline (TBS, Biorad)
- Mounting media ProLong gold with DAPI (Molecular Probes)
Next day: Module 2 begins!
Previous day: Query DNA repair capacity in tumor cells