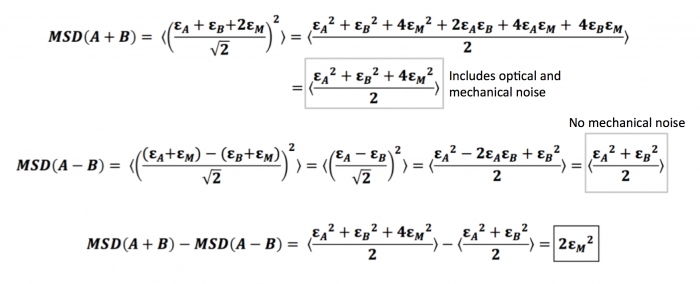Assignment 5, Part 1: MSD difference tracking and microscope stability
This is Part 1 of Assignment 5.
Overview
In the last assignment, you made movies of particles as theyjitterbugged and waltzed through your microscope's field of view. You developed code to track each particle's unique steps, and you computed each particle's mean squared displacement (MSD), diffusion coefficient, and the implied viscosity of the solutions the particles were suspended in. In this assignment, you'll be tracking particles again. But this time the particles are inside of cells. The cells have been cultured with 0.84 μm fluorescent microspheres, which enter the cells through the process of endocytosis. They are the same kind of cells you took pretty pictures of in assignment 3 (NIH 3T3). Trapped inside of the cells, the particles inside of cells will appear to be stationary to your naked eye. But they are definitely moving — albeit on a scale much smaller than the particles in glycerin solutions you measured previously.
How do we know whether our microscope is even capable of measuring such small motions? Could we only be measuring artifacts (like vibrations of the microscope, or drifting of the stage)?
By measuring the MSD of particles that have been glued down to a slide, we can actually diagnose what types of noise sources are contributing to our measurement, and thus what is the actual smallest motion (not due to noise) that we can measure. Some noise sources (like shot noise) are unavoidable, but others, (like mechanical vibrations and drift) we may be able to correct for.
MSD of sum and difference trajectories
When assessing the stability of one's microscope, it is helpful to look at the mean squared displacements (MSDs) of sum trajectories compared to those of difference trajectories.
Given two trajectories $ r_A $ and $ r_B $, the sum trajectory is given by $ r_A(t) + r_B(t) \over \sqrt 2 $, while the difference trajectory is given by $ r_A(t) - r_B(t) \over \sqrt 2 $
Here, we consider trajectory A, with its independent displacement $ \epsilon_A $, and trajectory B, with its independent displacement $ \epsilon_B $. Since these two trajectories come from particles that are glued well to the same slide (the slide you used for your stability measurements), they share the same mechanical noise that comes from vibrations and movements of the microscope stage. We will denote this mechanical noise as $ \epsilon_M $. Thus, the total error, or calculated displacement, from trajectory A would be $ \epsilon_A + \epsilon_M $, while that of trajectory B would similarly be $ \epsilon_B + \epsilon_M $. Note that we define displacement as the change in position over a time interval $ \tau $, as when we are calculating the MSD $ \left \langle {\left | r(t+\tau)-r(t) \right \vert}^2 \right \rangle $ . With this in mind, we can show that the difference between the two types of MSDs turns out to be two times the square of the mechanical noise as demonstrated below.
As you can see, measuring the MSD of the difference trajectories for two particles completely cancels out the mechanical noise. We can use the sum trajectories for two particles to diagnose the amount of mechanical noise initially present in your system: the difference between the MSDs of sum and difference trajectories is equal to (twice) the mechanical noise.
Develop sum and difference trajectory code
Modify your code from Assignment 4 to be able to track the sum and difference trajectories of particles in a movie.
Steve actually generously shared his code with you!
function [ DifferenceTrajectories, SumTrajectories ] = CalculateSumAndDifferenceTrajectories( Trajectories )
numberOfParticles = max( Trajectories(:,4) );
allCombinations = nchoosek( 1:numberOfParticles, 2 );
numberOfCombinations = size( allCombinations, 1 );
SumTrajectories = cell( numberOfCombinations, 1 );
DifferenceTrajectories = cell( numberOfCombinations, 1 );
virtualParticleNumber = 1;
for ii = 1:numberOfCombinations
firstParticleIndex = allCombinations( ii, 1 );
secondParticleIndex = allCombinations( ii, 2 );
firstTrajectory = Trajectories( Trajectories(:, 4) == firstParticleIndex, : );
secondTrajectory = Trajectories( Trajectories(:, 4) == secondParticleIndex, : );
framesInCommon = intersect( firstTrajectory(:, 3), secondTrajectory(:, 3 ) );
if( ~isempty( framesInCommon ) )
firstTrajectory = firstTrajectory(ismember(firstTrajectory(:, 3 ), framesInCommon), :);
secondTrajectory = secondTrajectory(ismember(secondTrajectory(:, 3 ), framesInCommon), :);
thisDifference = (firstTrajectory - secondTrajectory)/sqrt(2);
thisDifference(:,3) = framesInCommon';
thisDifference(:,4) = virtualParticleNumber;
DifferenceTrajectories{ii} = thisDifference;
thisSum = (firstTrajectory + secondTrajectory)/sqrt(2);
thisSum(:,3) = framesInCommon';
thisSum(:,4) = virtualParticleNumber;
SumTrajectories{ii} = thisSum;
virtualParticleNumber = virtualParticleNumber + 1;
end
end
DifferenceTrajectories = vertcat( DifferenceTrajectories{:} );
SumTrajectories = vertcat( SumTrajectories{:} );
end
Sp2020 Update:
The above code finds the sum and difference trajectories for every possible pairing of particles (given by this line of code:allCombinations = nchoosek( 1:numberOfParticles, 2 );). This is a bit overkill, especially when the number of particles is more than 5 or 6. Instead, you can use the code below that Steve wrote, which takes the sum and difference of particle pairs that are closest to each other. It's not perfect, since it discards a particle if there is an odd number of them, but it's an improvement on CalculateSumAndDifferenceTrajectories, above. This function also only keeps trajectories that exist for the entire length of the movie which is helpful for filtering out 'bad' particles.
If you have already completed assignment 5 using the old CalculateSumAndDifferenceTrajectories it's not a problem to keep using it, but know that you'll have many more traces on your MSD plot than necessary.
function [ DifferenceTrajectories, SumTrajectories ] = SumAndDifferenceTrajectoriesNearestNeighbor( Trajectories )
numberOfFrames = max(Trajectories(:,3));
[ counts, particleIds ] = groupcounts( Trajectories(:,4) );
particleIds( counts ~= numberOfFrames ) = [];
numberOfParticles = length( particleIds );
if( numberOfParticles < 2 )
DifferenceTrajectories = [];
SumTrajectories = [];
return
end
allPermutations = nchoosek( particleIds, numberOfParticles - mod( numberOfParticles, 2 ) );
numberOfPermutations = size( allPermutations, 1 );
initialPositions = Trajectories( Trajectories(:,3) == 1, : );
totalDistanceSquared = zeros( 1, numberOfPermutations );
for ii = 1:numberOfPermutations
for jj = 1:2:size( allPermutations, 2 )
firstParticlePosition = initialPositions(initialPositions(:,4) == allPermutations(ii,jj),1:2);
secondParticlePosition = initialPositions(initialPositions(:,4) == allPermutations(ii,jj+1));
totalDistanceSquared(ii) = totalDistanceSquared(ii) + sum( ( secondParticlePosition - firstParticlePosition ).^2 );
end
end
[~, minimumDistancePermutation ] = min( totalDistanceSquared );
bestPairing = allPermutations(minimumDistancePermutation,:);
allPairings = reshape( bestPairing, [], 2 );
numberOfCombinations = size( allPairings, 1 );
SumTrajectories = cell( numberOfCombinations, 1 );
DifferenceTrajectories = cell( numberOfCombinations, 1 );
virtualParticleNumber = 1;
for ii = 1:numberOfCombinations
firstParticleIndex = allPairings( ii, 1 );
secondParticleIndex = allPairings( ii, 2 );
firstTrajectory = Trajectories( Trajectories(:, 4) == firstParticleIndex, : );
secondTrajectory = Trajectories( Trajectories(:, 4) == secondParticleIndex, : );
framesInCommon = intersect( firstTrajectory(:, 3), secondTrajectory(:, 3 ) );
if( ~isempty( framesInCommon ) )
firstTrajectory = firstTrajectory(ismember(firstTrajectory(:, 3 ), framesInCommon), :);
secondTrajectory = secondTrajectory(ismember(secondTrajectory(:, 3 ), framesInCommon), :);
thisDifference = firstTrajectory - secondTrajectory;
thisDifference(:,3) = framesInCommon';
thisDifference(:,4) = virtualParticleNumber;
DifferenceTrajectories{ii} = thisDifference;
thisSum = firstTrajectory + secondTrajectory;
thisSum(:,3) = framesInCommon';
thisSum(:,4) = virtualParticleNumber;
SumTrajectories{ii} = thisSum;
virtualParticleNumber = virtualParticleNumber + 1;
end
end
DifferenceTrajectories = vertcat( DifferenceTrajectories{:} );
SumTrajectories = vertcat( SumTrajectories{:} );
Stability of microscope for particle tracking
Once you have developed and tested your code, use the following procedure to measure the location precision of your microscope:
- Bring a slide with fixed beads into focus.
- Choose a field of view in which you can see at least 3 beads using the 40× objective.
- Limit the field of view to only those beads by choosing a region of interest (ROI) in UsefulImageAcquisition tool. (Otherwise, you will have a gigantic movie that is hard to work with.)
- Acquire a movie of beads for 3 minutes at a frame rate of at least 10 fps.
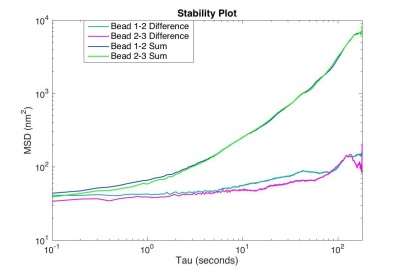
- Use the code you developed to track the particles, and calculate the sum and difference trajectories.
| |
Plot the MSD versus time interval of
|
The motion of particles endocytosed by 3T3 cells requires an extremely stable microscope, otherwise the real motion of the beads will be obscured by noise. Your microscope will be considered stable enough if the MSD for the difference trajectory of two fixed particles starts out less than 500 nm2 and remains less than 1000 nm2 up to t = 180 s. If the MSD is larger than that, refine your apparatus and methodology until you achieve that goal.
- Overview
- Part 1: MSD difference tracking and microscope stability
- Part 2: Live cell particle tracking of endocytosed beads
Back to 20.309 Main Page