20.109(S24):M1D6
Contents
Introduction
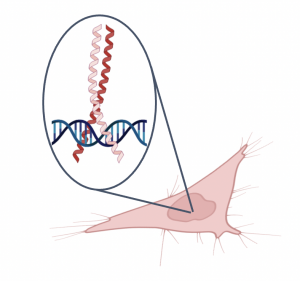
Today you will complete a nuclear extraction using the cells that you cultured in the previous laboratory session. Both the cell preparation and the nuclear extraction are steps that need to be completed for the EMSA experiment, which will examine the effect of the small molecules on Myc:MAX binding to a DNA target.In the EMSA experiment, we are evaluating binding in live cells treated with small molecule. Because we are using a cell-based version of this assay it is important to culture cells to a high density as this will ensure that there is enough Myc and MAX protein for visualization. In addition to seeding the cells, small molecule was added to the cell culture to test the effect on Myc:MAX binding to DNA. After the cell preparation, the nuclear contents of the cultured and treated cells must be extracted. As shown in the figure on the right Myc:MAX form a dimer that functions as a transcriptional regulator. This means that Myc:MAX will be located in the nucleus of the cell. To separate the nuclear proteins from the other cellular contents, a nuclear extraction protocol will be used as part of the the laboratory session today.
In a nuclear extraction, there are two lysis steps. For the first lysis step, cells are incubated for a brief time in a buffer solution that degrades the cellular membrane. The nuclei are then collected using centrifugation and incubated in a second lysis step. The second lysis step uses a different buffer solution and incubates for a longer period of time to degrade the nuclear membrane. After the nuclear membrane is degraded, the nuclear extract is collected.
Protocols
Part 1: Extract nuclear proteins from treated cell cultures
For timing reasons, the small molecule was incubated with your cell culture prior to the laboratory session. The small molecule was added at a final concentration of 30 μM and incubated with the cells for 24 hours.
- Obtain your cell culture from the front laboratory bench.
- Aspirate the media from the cell culture dish.
- Wash the cells by pippeting 5 mL of 1X PBS into the dish, then gently rock from side-to-side.
- Aspirate the 1X PBS from the cell culture dish.
- Prepare the Buffer I working reagent by mixing the following components:
- 100 μL of 10X Buffer I
- 900 μL of nuclease-free H2O
- 10 μL of DTT solution
- 10 μL of protease inhibitor
- Add all ~1 mL of Buffer I working reagent to the cell culture dish.
- Incubate cells for 10 minutes at 200 rpm in the cold room.
- The teaching assistant will take your cell culture to the cold room.
- Release the cells from the culture dish using a cell scraper.
- Be very careful at this step as it is easy to tip the dish and spill your cells!
- Transfer the cells to a microcentrifuge tube.
- Microcentrifuge the cells for 5 minutes at 12000 rpm at 4 °C.
- The teaching assistant will centrifuge your cells in the cold room.
- Carefully remove and discard the supernatent.
- Be sure to remove as much liquid as possible without disturbing the pellet.
- Keep your pellet on ice and prepare the Buffer II working reagent by mixing the following components:
- 50 μL of 5x Buffer II
- 200 μL of nuclease-free H2O
- 2.5 μL of DTT solution
- 2.5 μL of protease inhibitor
- Add all ~250 μL of Buffer II working reagent to the cell pellet.
- Do not tap or vortex the tube to resuspend the pellet! Instead, gently rock the tube until the pellet floats in the buffer.
- Incubate the pellet for 2 hours at 200 rpm in the cold room.
- The teaching assistant will take your cell culture to the cold room.
- Centrifuge the sample for 5 minutes at 12000 rpm at 4 °C.
- Carefully transfer the supernatent to a clean microcentrifuge tube.
- Label the tube with the supernatent as 'nuclear extract' and include your team information.
- The nuclear extract will be stored at -80 °C until the next laboratory session.
Part 2: Determine binding shifts from the DSF results
Today you will analyze the data for the DSF experiment. As a reminder DSF is a functional assay used to probe the interaction between a single protein of interest and a putative small molecule binder. In this assay, the melting temperature (Tm) of the protein is measured using a fluorescent dye and a change in the melting temperature (ΔTm) when the small molecule is present in the reaction is indicative of binding.
You will receive two XML files containing raw data from each well of the DSF assay plate over the specified range of temperatures. These files can be opened by dragging onto an open Excel window. The XML sheet with "Melt" in the file name will contain raw fluorescence intensity data, while the other sheet with "Tm" in its name will have the values for the first derivative of the melt curve. The leftmost column gives you the identity of the well. Refer back to your notes to determine what samples were loading in which wells. The “X” column denotes the temperature and the “Y” column denotes the fluorescence value for that well at that temperature.
One basic way to determine the "melting temperature," or Tm of the protein is to determine temperature at the inflection point of the melting curve. This inflection point would occur at the maximum value of the first derivative. The BioRad CFX machine we use actually exports the negative of the first derivative in the Excel file, so we will find the minimum value in the first derivative Excel file, and take the corresponding temperature to be the Tm in each condition.
- Open the Excel file corresponding to the first derivative data
- Copy over the column with the well identifier in a separate worksheet under column A, Copy over the temperature data in celsius in column B, and copy over the fluorescence data in column C
- At a row on the bottom of column C, type in the following command: =INDEX($B$FirstRow:$B$LastRow, MATCH(MIN(CFirstRow:CLastRow),CFirstRow:CLastRow,0)), where FirstRow corresponds to the row number of the first row containing data, and LastRow contains the row number of the last row containing data.
- Press enter, and double check that the listed temperature occurs at the minimum value of the first derivative.
- Then, drag the bottom left corner of the cell across all relevant columns to apply the formula to those columns of interest.
- Plot the columns relevant to your data set by making a scatter plot ("straight marked scatter"), having the temperature (values in column B) on the x-axis, and the first derivative values on the y-axis.
- Double check by eye that the values you calculated to be the melting temperatures correspond to the minimum values on the curves. (See example plot in the introduction section of the M2D5 wiki page)
- Next, you may also check to see what the melting curves look like in terms of raw fluorescence by plotting fluorescence intensity vs. temperature in the "Melt Curve RFU Results" file. Again, validate the results you found by eye to see if the Tm values correspond to the inflection point of the raw fluorescence melt curves.
- To determine whether ligand bound to either FKBP12 or FKBP35, plot the corresponding curves on the same plot as your DMSO control per compound tested. Quantify the shifts.
- Record the Tm values calculated for the small molecules you tested on the Class data page of the wiki.
In your laboratory notebook, complete the following:
- Record the Tm values.
- Attach the graphs generated as part of the analysis.
Reagent list
- Nuclear extraction kit (from Signosis)
- 10X Buffer I
- 5X Buffer II
- DTT solution
- protease inhibitor
- 1X PBS
- nuclease-free H2O
Next day: Complete EMSA experiment