Difference between revisions of "20.109:Module 3"
MAXINE JONAS (Talk | contribs) m (6 revisions: Test transfer 20.109(S07) to HostGator) |
|||
| (One intermediate revision by one user not shown) | |||
| Line 5: | Line 5: | ||
==Module 3== | ==Module 3== | ||
| − | '''Instructors:''' [ | + | '''Instructors:''' [http://openwetware.org/wiki/Natalie_Kuldell ] |
| − | '''TA:''' [ | + | '''TA:''' [http://openwetware.org/wiki/Andrea_Tentner ] |
| − | In this | + | Faced with the considerable challenge of packing more DNA into a cell, nature added proteins that reversibly compact the helix. The DNA can wind around these histone proteins, forming nucleosomes, that can then wind around each other to form chromatin. Other protein complexes modify and remodel the chromatin, making the DNA accessible for reading and copying. ���SAGA��� is a chromatin remodeling complexes in the model experimental yeast, S. cerevisiae, but it turns out not every protein in SAGA is needed for the yeast to survive. In this experiment we will delete one of the non-essential SAGA-subunits from the yeast genome and then ask how the resulting yeast, though alive, is affected. We will look for phenotypes that might indicate crippled functions and we will compare gene expression in the parent strain to each deletion strain using a microarray. Our individual experiments may identify targets for particular SAGA subunits while our class data may reveal genes that are commonly regulated by this remodeling complex. Given the structural information for SAGA that is recently available, we can hope to map our findings onto the complex and better understand the delicate balance between chromatin remodeling and gene expression. |
[[Image:Macintosh HD-Users-nkuldell-Desktop-ExpressionEng coverart S07.jpg|thumb|500 px|center| SAGA image from F.Winston<br> Mode of action model from P. Schultz <br> Mol Cell. 2004 Jul 23;15(2):199[[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=15260971&query_hl=4&itool=pubmed_docsum]]<br> Microarray image from N. Kuldell<br>]] | [[Image:Macintosh HD-Users-nkuldell-Desktop-ExpressionEng coverart S07.jpg|thumb|500 px|center| SAGA image from F.Winston<br> Mode of action model from P. Schultz <br> Mol Cell. 2004 Jul 23;15(2):199[[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=15260971&query_hl=4&itool=pubmed_docsum]]<br> Microarray image from N. Kuldell<br>]] | ||
| Line 16: | Line 16: | ||
[[20.109(S07): Yeast transformation| Module 3 Day 2: Yeast transformation]]<br> | [[20.109(S07): Yeast transformation| Module 3 Day 2: Yeast transformation]]<br> | ||
[[20.109(S07): Colony PCR| Module 3 Day 3: Colony PCR]] <br> | [[20.109(S07): Colony PCR| Module 3 Day 3: Colony PCR]] <br> | ||
| − | [ | + | [http://openwetware.org/wiki/20.109(S07):_Screen_for_phenotypes,_isolate_RNA | Module 3 Day 4: Screen for phenotypes, isolate RNA] <br> |
[[20.109(S07): cDNA synthesis and microarray| Module 3 Day 5: cDNA synthesis and microarray]] <br> | [[20.109(S07): cDNA synthesis and microarray| Module 3 Day 5: cDNA synthesis and microarray]] <br> | ||
[[20.109(S07): Microarray data analysis| Module 3 Day 6: Microarray data analysis]]<br> | [[20.109(S07): Microarray data analysis| Module 3 Day 6: Microarray data analysis]]<br> | ||
| − | [ | + | [http://openwetware.org/wiki/20.109(S07):_TA's_notes_for_module_3 | TA notes, mod 3] |
Latest revision as of 15:31, 15 June 2015
Module 3
Instructors: [2]
TA: [3]
Faced with the considerable challenge of packing more DNA into a cell, nature added proteins that reversibly compact the helix. The DNA can wind around these histone proteins, forming nucleosomes, that can then wind around each other to form chromatin. Other protein complexes modify and remodel the chromatin, making the DNA accessible for reading and copying. ���SAGA��� is a chromatin remodeling complexes in the model experimental yeast, S. cerevisiae, but it turns out not every protein in SAGA is needed for the yeast to survive. In this experiment we will delete one of the non-essential SAGA-subunits from the yeast genome and then ask how the resulting yeast, though alive, is affected. We will look for phenotypes that might indicate crippled functions and we will compare gene expression in the parent strain to each deletion strain using a microarray. Our individual experiments may identify targets for particular SAGA subunits while our class data may reveal genes that are commonly regulated by this remodeling complex. Given the structural information for SAGA that is recently available, we can hope to map our findings onto the complex and better understand the delicate balance between chromatin remodeling and gene expression.
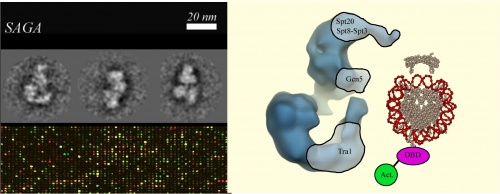
Mode of action model from P. Schultz
Mol Cell. 2004 Jul 23;15(2):199[[1]]
Microarray image from N. Kuldell
Module 3 Day 1: Start-up expression engineering
Module 3 Day 2: Yeast transformation
Module 3 Day 3: Colony PCR
| Module 3 Day 4: Screen for phenotypes, isolate RNA
Module 3 Day 5: cDNA synthesis and microarray
Module 3 Day 6: Microarray data analysis
