Difference between revisions of "20.109(S24):M1D6"
Noreen Lyell (Talk | contribs) (→Navigation links) |
Noreen Lyell (Talk | contribs) (→Protocols) |
||
| Line 35: | Line 35: | ||
*Attach the graphs generated as part of the analysis. | *Attach the graphs generated as part of the analysis. | ||
| + | ===Part 2: In-class paper discussion | ||
| + | To further help you in preparing your Data summary, we will discuss how similar data are presented in a publication from the Koehler laboratory. | ||
| + | Chen ''et al.'' titled "[[Media:SMMAlzABPeptide Koehler2010.pdf |Small molecule microarrays enable the discovery of compounds that bind the Alzheimer's Aβ peptide]]" (2010) ''J Am Chem Soc'' 132:17015-17022. | ||
| + | The initial experiment presented by Chen ''et. al.'' was an SMM that identified ligands binders of the amyloid-β (Aβ) petptide. This first step is very similar to what was done to identify the hits you are testing in this module! To further assess the results of the SMM, the authors completed several follow-up experiments to test the effect of the small molecule on functionality of the Aβ petptide. | ||
| + | |||
| + | In the context of your research, this article focuses on the next step experiments that can be performed after a drug candidate is discovered from a screen. Though you can use this article as guidance as you consider the experiments that could follow your screen, remember that the specific next step experiments should be related to the protein target and drug candidate(s) identified in your project. For this exercise, the focus in on how the data are organized and presented. | ||
| + | |||
| + | '''From the Introduction''' | ||
| + | |||
| + | Consider the key components of an introduction: | ||
| + | *What is the big picture? | ||
| + | *Is the importance of this research clear? | ||
| + | *Are you provided with the information you need to understand the research? | ||
| + | *Do the authors include a preview of the key results? | ||
| + | |||
| + | '''From the Results''' | ||
| + | |||
| + | Carefully examine the figures. First, read the captions and use the information to 'interpret' the data presented within the image. Second, read the text within the results section that describes the figure. | ||
| + | *Do you agree with the conclusion(s) reached by the authors? | ||
| + | *What controls were included and are they appropriate for the experiment performed? | ||
| + | *Are you convinced that the data are accurate and/or representative? | ||
| + | |||
| + | '''From the Discussion''' | ||
| + | |||
| + | Consider the following components of a discussion: | ||
| + | *Are the results summarized? | ||
| + | *Did the authors 'tie' the data together into a cohesive and well-interpreted story? | ||
| + | *Do the authors overreach when interpreting the data? | ||
| + | *Are the data linked back to the big picture from the introduction? | ||
| + | |||
| + | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
| + | *Based on your reading and the group discussion of the article, answer the questions above. | ||
==Navigation links== | ==Navigation links== | ||
Next day: [[20.109(S24):M1D7 |Perform electromobility shift assay (EMSA)]] <br> | Next day: [[20.109(S24):M1D7 |Perform electromobility shift assay (EMSA)]] <br> | ||
Previous day: [[20.109(S24):M1D5 |Prepare differential scanning flourimetry (DSF) experiment]] <br> | Previous day: [[20.109(S24):M1D5 |Prepare differential scanning flourimetry (DSF) experiment]] <br> | ||
Revision as of 17:44, 5 February 2024
Introduction
Today you will analyze the data for the DSF experiment. As a reminder DSF is a functional assay used to probe the interaction between a single protein of interest and a putative small molecule binder. In this assay, the melting temperature (Tm) of the protein is measured using a fluorescent dye and a change in the melting temperature (ΔTm) when the small molecule is present in the reaction is indicative of binding.
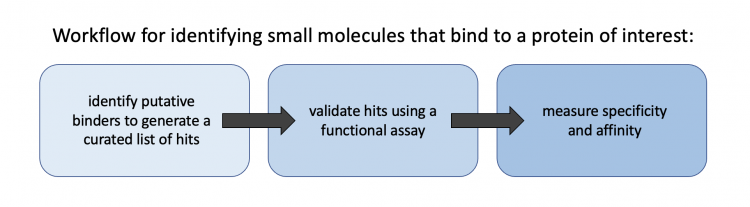
The DSF assay is one method used to validate 'hits' identified using a high-throughput approach, such as the SMM. Though an SMM was not used as part of this module, it was discussed to highlight how a researcher can easily screen through tens-of-thousands of compounds to generate a curated list of small molecules that can be further tested using a functional assay. For this module, we started with a short list of compounds based on known binders of our protein of interest. In either case, the workflow of this type of project is 1) identify putative binders using a screen or other method to generate a curated list of hits, 2) validate hits using a functional assay, 3) measure parameters such as specificity and affinity that exist between the protein of interest and validated hits.
Protocols
Part 1: Examine binding shifts
You will receive two XML files containing raw data from each well of a 384 well plate over the specified range of temperatures. These files can be opened by dragging onto an open Excel window. The XML sheet with "Melt" in the file name will contain raw fluorescence intensity data, while the other sheet with "Tm" in its name will have the values for the first derivative of the melt curve. The leftmost column gives you the identity of the well. Refer back to your notes to determine what samples were loading in which wells. The “X” column denotes the temperature and the “Y” column denotes the fluorescence value for that well at that temperature.
One basic way to determine the "melting temperature," or Tm of the protein is to determine temperature at the inflection point of the melting curve. This inflection point would occur at the maximum value of the first derivative. The BioRad CFX machine we use actually exports the negative of the first derivative in the Excel file, so we will find the minimum value in the first derivative Excel file, and take the corresponding temperature to be the Tm in each condition.
- Open the Excel file corresponding to the first derivative data
- Copy over the column with the well identifier in a separate worksheet under column A, Copy over the temperature data in celsius in column B, and copy over the fluorescence data in column C
- At a row on the bottom of column C, type in the following command: =INDEX($B$FirstRow:$B$LastRow, MATCH(MIN(CFirstRow:CLastRow),CFirstRow:CLastRow,0)), where FirstRow corresponds to the row number of the first row containing data, and LastRow contains the row number of the last row containing data.
- Press enter, and double check that the listed temperature occurs at the minimum value of the first derivative.
- Then, drag the bottom left corner of the cell across all relevant columns to apply the formula to those columns of interest.
- Plot the columns relevant to your data set by making a scatter plot ("straight marked scatter"), having the temperature (values in column B) on the x-axis, and the first derivative values on the y-axis.
- Double check by eye that the values you calculated to be the melting temperatures correspond to the minimum values on the curves. (See example plot in the introduction section of the M2D5 wiki page)
- Next, you may also check to see what the melting curves look like in terms of raw fluorescence by plotting fluorescence intensity vs. temperature in the "Melt Curve RFU Results" file. Again, validate the results you found by eye to see if the Tm values correspond to the inflection point of the raw fluorescence melt curves.
- To determine whether ligand bound to either FKBP12 or FKBP35, plot the corresponding curves on the same plot as your DMSO control per compound tested. Quantify the shifts.
- Record the Tm values calculated for the small molecules you tested on the Class data page of the wiki.
In your laboratory notebook, complete the following:
- Record the Tm values.
- Attach the graphs generated as part of the analysis.
===Part 2: In-class paper discussion
To further help you in preparing your Data summary, we will discuss how similar data are presented in a publication from the Koehler laboratory.
Chen et al. titled "Small molecule microarrays enable the discovery of compounds that bind the Alzheimer's Aβ peptide" (2010) J Am Chem Soc 132:17015-17022.
The initial experiment presented by Chen et. al. was an SMM that identified ligands binders of the amyloid-β (Aβ) petptide. This first step is very similar to what was done to identify the hits you are testing in this module! To further assess the results of the SMM, the authors completed several follow-up experiments to test the effect of the small molecule on functionality of the Aβ petptide.
In the context of your research, this article focuses on the next step experiments that can be performed after a drug candidate is discovered from a screen. Though you can use this article as guidance as you consider the experiments that could follow your screen, remember that the specific next step experiments should be related to the protein target and drug candidate(s) identified in your project. For this exercise, the focus in on how the data are organized and presented.
From the Introduction
Consider the key components of an introduction:
- What is the big picture?
- Is the importance of this research clear?
- Are you provided with the information you need to understand the research?
- Do the authors include a preview of the key results?
From the Results
Carefully examine the figures. First, read the captions and use the information to 'interpret' the data presented within the image. Second, read the text within the results section that describes the figure.
- Do you agree with the conclusion(s) reached by the authors?
- What controls were included and are they appropriate for the experiment performed?
- Are you convinced that the data are accurate and/or representative?
From the Discussion
Consider the following components of a discussion:
- Are the results summarized?
- Did the authors 'tie' the data together into a cohesive and well-interpreted story?
- Do the authors overreach when interpreting the data?
- Are the data linked back to the big picture from the introduction?
In your laboratory notebook, complete the following:
- Based on your reading and the group discussion of the article, answer the questions above.
Next day: Perform electromobility shift assay (EMSA)