Difference between revisions of "20.109(S23):M1D8"
Noreen Lyell (Talk | contribs) (Created page with "<div style="padding: 10px; width: 820px; border: 5px solid #434a43;"> {{Template:20.109(S23)}} ==Introduction== ==Protocols== ==Reagents list== ==Navigation links== Nex...") |
Noreen Lyell (Talk | contribs) (→Protocols) |
||
| Line 7: | Line 7: | ||
==Protocols== | ==Protocols== | ||
| + | |||
| + | |||
| + | ====Identify common features in hits==== | ||
| + | |||
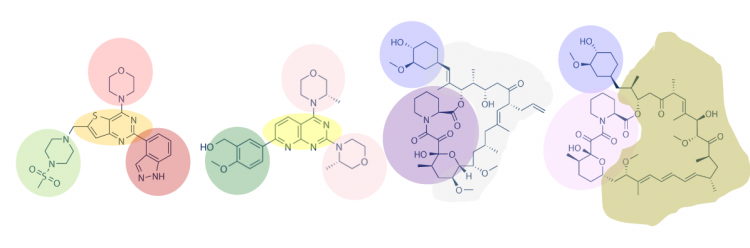
| + | One method for assessing protein-small molecule binding is to visually inspect known small molecule binders for common features / structures. To do this you will carefully examine the hits and identify any common features / structures. As in the image below, it is possible that multiple features will be present within the same small molecule. | ||
| + | |||
| + | [[Image:Sp17 20.109 M1D7 chemical structure features.png|thumb|750px|center|]] | ||
| + | |||
| + | |||
| + | Review the hits that were identified in the SMM screen completed for TDP43-RRM12. To see the chemical structures, translate the SMILES strings using one of the methods described in the text below the table. It may be easier to copy / paste the small molecule images into a powerpoint file so you can readily see all of the structures. Also, it may be helpful to use a color-coding system (like the one in the image provided above) to highlight features / structures that are common to the hits. | ||
| + | |||
| + | '''These online resources may be helpful to learning more about the hits identified in the SMM:''' | ||
| + | *Cloud version of ChemDraw [https://chemdrawdirect.perkinelmer.cloud/js/sample/index.html here]. | ||
| + | **Copy and paste the small molecule smiles into the work space to get a chemical structure | ||
| + | *Platform to transform the smiles information into a PubChem ID [https://pubchem.ncbi.nlm.nih.gov/idexchange/idexchange.cgi here]. | ||
| + | **Copy and paste the smiles into the input ID search to determine the ID number. | ||
| + | *PubChem database of chemical information [https://pubchem.ncbi.nlm.nih.gov here]. | ||
| + | **Includes small molecule molecular weight and other useful information. | ||
| + | |||
| + | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
| + | |||
| + | *How many features did you identify that are present in two or more of the small molecules that putatively bind TDP43-RRM12? Are there more or less than you expected? | ||
| + | *Is there a feature present in all of the identified small molecules? What might this suggest about the binding site(s) and / or binding ability of TDP43-RRM12? | ||
| + | *Can you assign the identified small molecules to sub-groups based on the common features that are present? | ||
| + | *What might the different features represent? More specifically, consider whether each subgroup has a unique binding site on the target protein or if each subgroup represents different solutions for interacting with the same binding site. | ||
| + | *How might you make modifications to the small molecules / features to probe binding? As a hint, consider how different functional groups could be positioned at a given site without altering qualitative binding in the SMM assay to translate that into some testable ideas. | ||
==Reagents list== | ==Reagents list== | ||
Revision as of 20:37, 7 February 2023
Contents
Introduction
Protocols
Identify common features in hits
One method for assessing protein-small molecule binding is to visually inspect known small molecule binders for common features / structures. To do this you will carefully examine the hits and identify any common features / structures. As in the image below, it is possible that multiple features will be present within the same small molecule.
Review the hits that were identified in the SMM screen completed for TDP43-RRM12. To see the chemical structures, translate the SMILES strings using one of the methods described in the text below the table. It may be easier to copy / paste the small molecule images into a powerpoint file so you can readily see all of the structures. Also, it may be helpful to use a color-coding system (like the one in the image provided above) to highlight features / structures that are common to the hits.
These online resources may be helpful to learning more about the hits identified in the SMM:
- Cloud version of ChemDraw here.
- Copy and paste the small molecule smiles into the work space to get a chemical structure
- Platform to transform the smiles information into a PubChem ID here.
- Copy and paste the smiles into the input ID search to determine the ID number.
- PubChem database of chemical information here.
- Includes small molecule molecular weight and other useful information.
In your laboratory notebook, complete the following:
- How many features did you identify that are present in two or more of the small molecules that putatively bind TDP43-RRM12? Are there more or less than you expected?
- Is there a feature present in all of the identified small molecules? What might this suggest about the binding site(s) and / or binding ability of TDP43-RRM12?
- Can you assign the identified small molecules to sub-groups based on the common features that are present?
- What might the different features represent? More specifically, consider whether each subgroup has a unique binding site on the target protein or if each subgroup represents different solutions for interacting with the same binding site.
- How might you make modifications to the small molecules / features to probe binding? As a hint, consider how different functional groups could be positioned at a given site without altering qualitative binding in the SMM assay to translate that into some testable ideas.
Reagents list
Next day: Determine mutagenesis strategy