Difference between revisions of "20.109(S21):M1D4"
(→Part 3:Applying our knowledge of immunity to pandemic problem solving) |
|||
| (61 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | <div style="padding: 10px; width: 820px; border: 5px solid # | + | <div style="padding: 10px; width: 820px; border: 5px solid #1e0c9f;"> |
{{Template:20.109(S21)}} | {{Template:20.109(S21)}} | ||
| − | ==Introduction: | + | ==Introduction:Purify and sequence clone plasmids== |
| − | Today you will submit | + | Today you will purify the plasmid DNA from ''E. coli'' and submit for sequencing analysis. We want to determine the sequence differences between these "mutant" clones and our characterized parental scFv clone. These newly isolated plasmids encode for unique single-chain antibody fragment that may show improved binding to our antigen, lysozyme. |
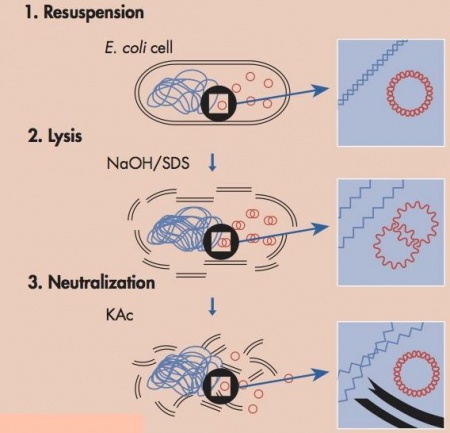
| − | + | [[Image:Qiagen_alkalinelysis.jpg|thumb|right|450px|'''Schematic of alkaline lysis: Blue DNA genomic and red DNA plasmid. Image by Qiagen''']] Today we will harvest plasmid DNA from ''E. coli'' using a purification method called alkaline lysis. In the last lab section, for yeast there needs to be one additional step to help break down the cell wall, but overall goal of each step is the same between both purification protocols--separate the plasmid DNA from the chromosomal DNA and cellular debris, allowing the plasmid DNA to be studied further. '''The key difference between plasmid DNA and chromosomal DNA is size and this difference is what is used to separate the two components.''' | |
| − | + | In this protocol the media is removed from the cells by centrifugation. The cells are '''first resuspended''' in a solution that contains Tris, to buffer the cells, and EDTA to bind divalent cations in the lipid bilayer, thereby weakening the cell envelope. '''Second an alkaline (basic) lysis buffer''' is added that contains sodium hydroxide and the detergent sodium dodecyl sulfate (SDS). The base denatures the cell’s DNA, both chromosomal and plasmid, while the detergent dissolves the cellular proteins and lipids. '''Third the pH of the solution is returned to neutral''' by adding a mixture of acetic acid and potassium acetate. At neutral pH the SDS precipitates from solution, carrying with it the dissolved proteins and lipids. The DNA strands renature at neutral pH. The chromosomal DNA, which is much longer than the plasmid DNA, renatures as a tangle that gets trapped in the SDS precipitate. The smaller plasmid DNA renatures normally and stays in solution, effectively separating plasmid DNA from the chromosomal DNA and the proteins and lipids of the cell. At this point the solution is spun at a high speed and soluble fraction, including the plasmid, is kept for further purification and the insoluble fraction, the macromolecules and chromosomal DNA is pelleted and thrown away. Following these steps there are several more washes to purify the plasmid DNA but the major purification work, separating the plasmid from the chromosomal DNA and cell lysate, is completed by the three steps shown in the figure above. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
==Protocols== | ==Protocols== | ||
| − | To use | + | === Part 1: Harvest plasmid (Miniprep) from ''E. coli'' === |
| + | '''Motivation:''' To analyze and further use the plasmids containing unique scFv genes, the DNA must be harvested from E. coli. | ||
| + | |||
| + | Since the last lab session, the transformed ''E. coli'' bacteria grew on LB agar plated with ampicillin antibiotic selection overnight at 37°C. Only bacteria that took up plasmid with an ampicillin selection marker during the heat shock were able to grow, the non-transformants died. The next day visible colonies were picked off the LB+amp plates and grown in LB+amp liquid culture overnight. Growing the bacteria in liquid culture allows one to easily harvest plasmid from a high number of cells. | ||
| + | Note: The procedure for DNA isolation at this scale (1-2mL) is commonly termed "mini-prep," which distinguishes it from a “maxi-prep” that involves a larger volume (100mL) of cells and additional steps of purification. | ||
| + | |||
| + | #Pick up your two cultures, which are growing in test tubes labeled with your team color. Label two 1.5mL tubes to reflect your samples (scFv_clone#1 and #2). | ||
| + | #Vortex the bacteria and pour ~1.5 mL of each candidate into a 1.5mL tube. | ||
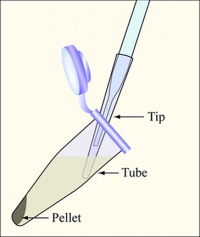
| + | #Balance the tubes in the microfuge, spin them at maximum speed for 2 min, and remove the supernatants with the vacuum aspirator. [[Image:Removing cells.jpg|thumb|right|200px|'''Diagram showing how to aspirate the supernatant.''' Be careful to remove as few cells as possible.]] | ||
| + | #Resuspend each cell pellet in 250 μL buffer P1. | ||
| + | #*Buffer P1 contains RNase so that we collect only our nucleic acid of interest, DNA. | ||
| + | #Add 250 μL of buffer P2 to each tube, and mix by inversion until the suspension is homogeneous. About 4-6 inversions of the tube should suffice. You may incubate here for '''up to 5 minutes, but not more'''. | ||
| + | #*Buffer P2 contains sodium hydroxide and SDS for alkaline lysis. | ||
| + | #Add 350 μL buffer N3 to each tube, and mix '''immediately''' by inversion (4-10 times). | ||
| + | #*Buffer N3 contains acetic acid which neutralizes the pH. The plasmid DNA renatures while chromosomal DNA / proteins / lipids precipitate into visible white aggregates. | ||
| + | #Centrifuge for 10 minutes at maximum speed. Note that you will be saving the '''supernatant''' after this step. | ||
| + | #*Meanwhile, prepare 2 labeled QIAprep columns, one for each candidate clone. You must add the label directly to the blue column and not the clear tube. | ||
| + | #Transfer the entire supernatant to the column and centrifuge for 1 min. Discard the eluant into a tube labeled ''''Qiagen waste'.''' | ||
| + | #Add 0.5 mL PB to each column, then spin for 1 min and discard the eluant into the Qiagen waste. | ||
| + | #Next wash with 0.75 mL PE, with a 1 min spin step as usual. Discard the ethanol in the Qiagen waste. | ||
| + | #After removing the PE, spin the mostly dry column for 1 more minute. | ||
| + | #*It is important to remove all traces of ethanol, as they may interfere with subsequent work with the DNA. | ||
| + | #Transfer each column insert (blue) to a 1.5mL tube with the lid cut off. | ||
| + | #*Use the scissors in your drawer to cut the lids off two tubes. | ||
| + | #Add 30 μL of distilled H<sub>2</sub>O pH ~8 to the top center of the column, wait 1 min, and then spin 1 min to elute your DNA. | ||
| + | #Cap the trimmed tube or transfer elution to new 1.5mL tube. | ||
| + | #You will now take your samples to the Nanodrop to measure the DNA's absorbance to quantify the concentration. | ||
| + | |||
| + | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
| + | #Describe where the plasmid DNA is in the following steps: | ||
| + | #*In step three when bacteria is pelleted, where is plasmid DNA? | ||
| + | #*In step four when bacteria is resuspended in P1, where is plasmid DNA? | ||
| + | #*In step seven after the 10min spin, is the plasmid DNA in solid precipitate or liquid supernatant solution? | ||
| + | #*In step nine when the column in washed with buffer PB, where is plasmid DNA? | ||
| + | #*In step thirteen after the 1min spin, is the plasmid DNA in the column or flow through? | ||
| + | |||
| + | === Part 2: Sequencing scFv clones === | ||
| + | '''Motivation:''' To determine the mutations in the scFv DNA sequence the plasmid DNA is prepared and submitted for Sanger sequencing. | ||
| + | |||
| + | Just as PCR amplification reactions require a primer for initiation, primers are also needed for sequencing reactions. Distinguishable sequence readout typically begins about 40-50 bases downstream of the primer binding site, and continues for ~1000 bases at most. Thus, usually forward and reverse primers must be used to fully view genes > 1 kb in size. The target sequence for your scFv is shorter than 1000 bp, but we will sequence with both a forward and reverse primer to double-check that we have an accurate representation of the clone sequence. | ||
| + | |||
| + | The primers you will use today are below: | ||
| + | |||
| + | <center> | ||
| + | {| border="1" | ||
| + | ! Primer | ||
| + | ! Sequence | ||
| + | |- | ||
| + | | scFv forward primer | ||
| + | | 5' - GTTCCAGACTACGCTCTGCAGG- 3' | ||
| + | |- | ||
| + | | scFv reverse primer | ||
| + | | 5' - GATTTTGTTACATCTACACTGTTG- 3' | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| + | |||
| + | The recommended composition of sequencing reactions is ~800 ng of plasmid DNA and 25 pmoles of one sequencing primer in a final volume of 15 μL. The miniprep'd plasmid should have ~200 ng of nucleic acid/μL, so we will estimate the amount appropriate for our reactions. | ||
| + | |||
| + | Because you will examine the sequence of your potential plasmids using both a forward and a reverse primer, '''you will need to prepare two reactions for each clone'''. Thus you will have a total of four sequencing reactions. For each reaction, combine the following reagents directly in the appropriate tube within the PCR-tube strip, as noted in the table below: | ||
| + | # 6 μL nuclease-free water | ||
| + | # 4 μL of your plasmid DNA candidate | ||
| + | # 5 μL of the primer stock on the teaching bench (the stock concentration is 5 pmol/μL) | ||
| + | #* Please add the forward primer to the odd numbered tubes and the reverse primer to the even numbered tubes (''i.e.'' tube #1 contains scFv#1 plasmid DNA and forward primer, tube #2 contains scFv#1 plasmid DNA and reverse primer, etc). | ||
| + | |||
| + | The side of each tube is numerically labeled and you should use only the four tubes assigned to your group. The teaching faculty submit the samples to the Genewiz company for sequencing via the drop-off box in the hall. | ||
| + | |||
| + | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following:<br> | ||
| + | #Open the Lyso_scFv_6 Snapgene file you started M1D1 or open this [[Media:Sp21_Lyso_scFv_6.dna|Sequence file]]. | ||
| + | #*If you need to review basic tools in SnapGene remember our exercise on [http://engineerbiology.org/wiki/20.109(S21):M1D1 M1D1]. | ||
| + | #Find and copy the forward and reverse sequencing primers in Part 2 on this page. Add them to the sequence file and determine the size of the sequencing product. | ||
| + | #*Edit->Find->Find DNA sequence then Primer-> Add Primer | ||
| + | #Do you predict you will get high quality forward and reverse sequence coverage of the possible mutations created M1D1? Why or why not? | ||
| + | |||
| + | === Part 3:Applying our knowledge of immunity to pandemic problem solving=== | ||
| + | '''Motivation:''' Scientific knowledge doesn't exist in a vacuum. As biological engineers and science communicators you'll need to integrate your technical knowledge with ethical issues that impact society. Today we'll learn more about immunity in the context of herd immunity and have a group discussion about the logic and ethics of Massachusetts vaccine rollout policies. | ||
| + | |||
| + | This group discussion has been adapted from "It takes a herd: How can we use immunity to combat an emerging infectious disease?" By Adam J. Kleinschmit at the Department of Natural and Applied Sciences, University of Dubuque, Dubuque IA. | ||
| + | |||
| + | '''Round 1''': Individual Research <br> | ||
| + | Each person will be assigned short background reading in one of four subjects relating to COVID-19: (1) Reaching herd immunity, (2) acquired active immunity through vaccination, (3) naturally acquired immunity by infection, and (4) acquired passive immunity through the creation of therapeutic monoclonal antibody cocktails. You'll each be assigned a subgroup topic with 3 articles. Please read the first article then review the next two if you have time. <br> | ||
| − | + | First review information we discussed in lecture today via this [[media: Sp21_Herd_background.pdf |primer on immunity and herd immunity]]. | |
| + | *Red and Orange team, Herd immunity: [https://www.opb.org/article/2021/02/24/getting-back-to-normal-in-oregon-means-getting-to-covid-19-herd-immunity/ article 1], [https://www.who.int/news-room/q-a-detail/herd-immunity-lockdowns-and-covid-19 article 2], and [[media: We’ll_Have_Herd_Immunity_by_April_-_WSJ.pdf |article 3]] | ||
| + | *Yellow and Green team, vaccination: [https://www.nytimes.com/interactive/2021/health/how-covid-19-vaccines-work.html article 1], [https://www.healthline.com/health-news/how-much-will-it-cost-to-get-a-covid-19-vaccine article 2], and [https://www.nature.com/articles/d41586-021-00396-2 article 3]. | ||
| + | *Blue and Pink team, immunity through infection: [https://directorsblog.nih.gov/2020/12/08/study-of-healthcare-workers-shows-covid-19-immunity-lasts-many-months/ article1], [https://jamanetwork.com/journals/jamainternalmedicine/fullarticle/2773575 article 2], and [https://www.latimes.com/science/story/2021-02-24/coronavirus-infection-leads-to-immunity-thats-comparable-to-a-covid-19-vaccine article 3]. | ||
| + | *Purple and Teal team, therapeutic antibodies: [https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibody-treatment-covid-19 article 1], [https://www.nytimes.com/2021/02/09/world/covid-coronavirus-lilly-antibody-treatment.html article 2], and [https://www.npr.org/sections/health-shots/2020/10/28/928841997/government-signs-deal-for-covid-19-treatments-from-eli-lilly article 3]. | ||
| + | '''Round 2''': Debrief Groups <br> | ||
| + | Debrief groups will be assembled so people can share their expertise. Each group will contain at least one person from the four focus groups in round 1 and will be placed in a breakout together. Discuss the topics you were assigned in the context of moving quickly toward COVID-19 herd immunity. You'll take group notes via the google doc [https://docs.google.com/document/d/1_u46hGS7005dppB-BBkgOFk2GbtEdbeOZZU8fk-XKOA/edit?usp=sharing here]. Please address the prompts and include your groups thoughts. | ||
| − | + | '''Round 3''': Whole Group Discussion <br> | |
| − | + | Using our new insights and knowledge of herd immunity, we will discuss recent Massachusetts State policies that are designed to quickly reach herd immunity while providing vaccine rollout in an ethical way. <br> | |
| − | + | Policy 1: Effective Thursday, February 11th, an individual who accompanies a person age 75 or older to one of four mass vaccination sites to receive the vaccine will be eligible to receive the vaccine too, if they have an appointment booked. Both the companion and the individual age 75 or older must have an appointment for the same day and both individuals must be present. Only one companion is permitted. The companion must attest that they are accompanying the individual to the appointment. <br> | |
| + | See the [[media: COVID19_Vaccine_75_Companion_Information_2.10.2021_FINAL_.docx|state announcement]] here. | ||
| − | + | Policy 2:Effective Feb. 17, 2021 Massachusetts will be focusing its upcoming vaccine distribution efforts on its mass vaccination sites, such as Gillette Stadium and Fenway Park, and its partnerships with retail drugstores CVS and Walgreens. This means that venues such as MIT Medical are unlikely to receive vaccine in the coming weeks. See the announcement [https://now.mit.edu/latest-updates/next-step-in-states-vaccine-rollout-plan-starts-feb-18/ here]. | |
| − | + | <br> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | Policy 3:Feb. 12th, Nearly every member of the Massachusetts congressional delegation is calling on Gov. Charlie Baker to set up a system to allow residents to pre-register and receive a notification for COVID-19 vaccine appointments.See news article [https://www.boston.com/news/coronavirus/2021/02/17/charlie-baker-responds-mass-delegation-vaccine-preregistration here] and copy of the letter [[media: Gov-Baker-letter-Vaccines-2-12-21-FINAL2-602ae06be57a3.pdf| here]]. | |
| + | <br> | ||
| − | + | *Is the policy ethical? | |
| + | *Is the policy a logical decision to get Massachusetts to herd immunity faster? | ||
| + | *Are there better approaches to this problem? | ||
| − | + | Additional reading: | |
| − | + | *[https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibodies-treatment-covid-19 Antibody based COVID-19 treatments] | |
| − | + | *[https://www.nytimes.com/2020/12/23/health/coronavirus-antibody-drugs.html Cost of antibody based drugs] | |
| − | + | *[https://www.cdc.gov/coronavirus/2019-ncov/vaccines/facts.html CDC vaccine facts] | |
| − | + | *[https://www.cnet.com/health/covid-19-vaccine-employer-requirements-hidden-costs-when-youll-get-vaccinated-more/ Ethical dilemmas of COVID-19 vaccination] | |
| − | + | *[https://www.vumc.org/viiii/immuknow/debating-duration-covid-19-immunity Debating duration of immunity] | |
| − | + | *[https://www.npr.org/sections/health-shots/2021/02/03/963373971/a-rocky-road-on-the-way-to-herd-immunity-for-covid-19 Rocky road to herd immunity] | |
| − | + | *[[media: Massachusetts hospitals to resume offering COVID-19 vaccine - The Boston Globe.pdf | Choice to favor mass vaccination sites over hospitals]] | |
| − | + | *[[media: Baker details $4.7m vaccine equity initiative, says 50,000 new vaccine appointments will go live Thursday - The Boston Globe.pdf | Massachusetts $4.7m vaccine equity initiative]] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | ==Reagents== | ||
| + | *Chemically competent ''E. coli'' NEB5a (genotype: fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17'') | ||
| + | *SOC medium: 2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose | ||
| + | *LB+Amp plates | ||
| + | **Luria-Bertani (LB) broth: 1% tryptone, 0.5% yeast extract, and 1% NaCl | ||
| + | **Plates prepared by adding 1.5% agar and 100 μg/mL ampicillin (Amp) to LB | ||
| + | *QIAprep Spin Miniprep Kit (from Qiagen) | ||
| + | **buffer P1 | ||
| + | **buffer P2 | ||
| + | **buffer N3 | ||
| + | **buffer PB | ||
| + | **buffer PE | ||
==Reagents list== | ==Reagents list== | ||
| Line 68: | Line 145: | ||
==Navigation links== | ==Navigation links== | ||
| − | Next day: [[]]<br> | + | Next day: [[20.109(S21):M1D5 | Analyze clone sequences and choose clone to characterize ]]<br> |
| − | Previous day: [[ ]] | + | Previous day: [[20.109(S21):M1D3 | Harvest clone plasmids from library ]] |
| + | [[20.109(S21):Homework#Due_M1D5 | Homework due M1D5]] | ||
Latest revision as of 14:37, 3 March 2021
Contents
Introduction:Purify and sequence clone plasmids
Today you will purify the plasmid DNA from E. coli and submit for sequencing analysis. We want to determine the sequence differences between these "mutant" clones and our characterized parental scFv clone. These newly isolated plasmids encode for unique single-chain antibody fragment that may show improved binding to our antigen, lysozyme.
Today we will harvest plasmid DNA from E. coli using a purification method called alkaline lysis. In the last lab section, for yeast there needs to be one additional step to help break down the cell wall, but overall goal of each step is the same between both purification protocols--separate the plasmid DNA from the chromosomal DNA and cellular debris, allowing the plasmid DNA to be studied further. The key difference between plasmid DNA and chromosomal DNA is size and this difference is what is used to separate the two components.In this protocol the media is removed from the cells by centrifugation. The cells are first resuspended in a solution that contains Tris, to buffer the cells, and EDTA to bind divalent cations in the lipid bilayer, thereby weakening the cell envelope. Second an alkaline (basic) lysis buffer is added that contains sodium hydroxide and the detergent sodium dodecyl sulfate (SDS). The base denatures the cell’s DNA, both chromosomal and plasmid, while the detergent dissolves the cellular proteins and lipids. Third the pH of the solution is returned to neutral by adding a mixture of acetic acid and potassium acetate. At neutral pH the SDS precipitates from solution, carrying with it the dissolved proteins and lipids. The DNA strands renature at neutral pH. The chromosomal DNA, which is much longer than the plasmid DNA, renatures as a tangle that gets trapped in the SDS precipitate. The smaller plasmid DNA renatures normally and stays in solution, effectively separating plasmid DNA from the chromosomal DNA and the proteins and lipids of the cell. At this point the solution is spun at a high speed and soluble fraction, including the plasmid, is kept for further purification and the insoluble fraction, the macromolecules and chromosomal DNA is pelleted and thrown away. Following these steps there are several more washes to purify the plasmid DNA but the major purification work, separating the plasmid from the chromosomal DNA and cell lysate, is completed by the three steps shown in the figure above.
Protocols
Part 1: Harvest plasmid (Miniprep) from E. coli
Motivation: To analyze and further use the plasmids containing unique scFv genes, the DNA must be harvested from E. coli.
Since the last lab session, the transformed E. coli bacteria grew on LB agar plated with ampicillin antibiotic selection overnight at 37°C. Only bacteria that took up plasmid with an ampicillin selection marker during the heat shock were able to grow, the non-transformants died. The next day visible colonies were picked off the LB+amp plates and grown in LB+amp liquid culture overnight. Growing the bacteria in liquid culture allows one to easily harvest plasmid from a high number of cells. Note: The procedure for DNA isolation at this scale (1-2mL) is commonly termed "mini-prep," which distinguishes it from a “maxi-prep” that involves a larger volume (100mL) of cells and additional steps of purification.
- Pick up your two cultures, which are growing in test tubes labeled with your team color. Label two 1.5mL tubes to reflect your samples (scFv_clone#1 and #2).
- Vortex the bacteria and pour ~1.5 mL of each candidate into a 1.5mL tube.
- Balance the tubes in the microfuge, spin them at maximum speed for 2 min, and remove the supernatants with the vacuum aspirator.
- Resuspend each cell pellet in 250 μL buffer P1.
- Buffer P1 contains RNase so that we collect only our nucleic acid of interest, DNA.
- Add 250 μL of buffer P2 to each tube, and mix by inversion until the suspension is homogeneous. About 4-6 inversions of the tube should suffice. You may incubate here for up to 5 minutes, but not more.
- Buffer P2 contains sodium hydroxide and SDS for alkaline lysis.
- Add 350 μL buffer N3 to each tube, and mix immediately by inversion (4-10 times).
- Buffer N3 contains acetic acid which neutralizes the pH. The plasmid DNA renatures while chromosomal DNA / proteins / lipids precipitate into visible white aggregates.
- Centrifuge for 10 minutes at maximum speed. Note that you will be saving the supernatant after this step.
- Meanwhile, prepare 2 labeled QIAprep columns, one for each candidate clone. You must add the label directly to the blue column and not the clear tube.
- Transfer the entire supernatant to the column and centrifuge for 1 min. Discard the eluant into a tube labeled 'Qiagen waste'.
- Add 0.5 mL PB to each column, then spin for 1 min and discard the eluant into the Qiagen waste.
- Next wash with 0.75 mL PE, with a 1 min spin step as usual. Discard the ethanol in the Qiagen waste.
- After removing the PE, spin the mostly dry column for 1 more minute.
- It is important to remove all traces of ethanol, as they may interfere with subsequent work with the DNA.
- Transfer each column insert (blue) to a 1.5mL tube with the lid cut off.
- Use the scissors in your drawer to cut the lids off two tubes.
- Add 30 μL of distilled H2O pH ~8 to the top center of the column, wait 1 min, and then spin 1 min to elute your DNA.
- Cap the trimmed tube or transfer elution to new 1.5mL tube.
- You will now take your samples to the Nanodrop to measure the DNA's absorbance to quantify the concentration.
In your laboratory notebook, complete the following:
- Describe where the plasmid DNA is in the following steps:
- In step three when bacteria is pelleted, where is plasmid DNA?
- In step four when bacteria is resuspended in P1, where is plasmid DNA?
- In step seven after the 10min spin, is the plasmid DNA in solid precipitate or liquid supernatant solution?
- In step nine when the column in washed with buffer PB, where is plasmid DNA?
- In step thirteen after the 1min spin, is the plasmid DNA in the column or flow through?
Part 2: Sequencing scFv clones
Motivation: To determine the mutations in the scFv DNA sequence the plasmid DNA is prepared and submitted for Sanger sequencing.
Just as PCR amplification reactions require a primer for initiation, primers are also needed for sequencing reactions. Distinguishable sequence readout typically begins about 40-50 bases downstream of the primer binding site, and continues for ~1000 bases at most. Thus, usually forward and reverse primers must be used to fully view genes > 1 kb in size. The target sequence for your scFv is shorter than 1000 bp, but we will sequence with both a forward and reverse primer to double-check that we have an accurate representation of the clone sequence.
The primers you will use today are below:
| Primer | Sequence |
|---|---|
| scFv forward primer | 5' - GTTCCAGACTACGCTCTGCAGG- 3' |
| scFv reverse primer | 5' - GATTTTGTTACATCTACACTGTTG- 3' |
The recommended composition of sequencing reactions is ~800 ng of plasmid DNA and 25 pmoles of one sequencing primer in a final volume of 15 μL. The miniprep'd plasmid should have ~200 ng of nucleic acid/μL, so we will estimate the amount appropriate for our reactions.
Because you will examine the sequence of your potential plasmids using both a forward and a reverse primer, you will need to prepare two reactions for each clone. Thus you will have a total of four sequencing reactions. For each reaction, combine the following reagents directly in the appropriate tube within the PCR-tube strip, as noted in the table below:
- 6 μL nuclease-free water
- 4 μL of your plasmid DNA candidate
- 5 μL of the primer stock on the teaching bench (the stock concentration is 5 pmol/μL)
- Please add the forward primer to the odd numbered tubes and the reverse primer to the even numbered tubes (i.e. tube #1 contains scFv#1 plasmid DNA and forward primer, tube #2 contains scFv#1 plasmid DNA and reverse primer, etc).
The side of each tube is numerically labeled and you should use only the four tubes assigned to your group. The teaching faculty submit the samples to the Genewiz company for sequencing via the drop-off box in the hall.
In your laboratory notebook, complete the following:
- Open the Lyso_scFv_6 Snapgene file you started M1D1 or open this Sequence file.
- If you need to review basic tools in SnapGene remember our exercise on M1D1.
- Find and copy the forward and reverse sequencing primers in Part 2 on this page. Add them to the sequence file and determine the size of the sequencing product.
- Edit->Find->Find DNA sequence then Primer-> Add Primer
- Do you predict you will get high quality forward and reverse sequence coverage of the possible mutations created M1D1? Why or why not?
Part 3:Applying our knowledge of immunity to pandemic problem solving
Motivation: Scientific knowledge doesn't exist in a vacuum. As biological engineers and science communicators you'll need to integrate your technical knowledge with ethical issues that impact society. Today we'll learn more about immunity in the context of herd immunity and have a group discussion about the logic and ethics of Massachusetts vaccine rollout policies.
This group discussion has been adapted from "It takes a herd: How can we use immunity to combat an emerging infectious disease?" By Adam J. Kleinschmit at the Department of Natural and Applied Sciences, University of Dubuque, Dubuque IA.
Round 1: Individual Research
Each person will be assigned short background reading in one of four subjects relating to COVID-19: (1) Reaching herd immunity, (2) acquired active immunity through vaccination, (3) naturally acquired immunity by infection, and (4) acquired passive immunity through the creation of therapeutic monoclonal antibody cocktails. You'll each be assigned a subgroup topic with 3 articles. Please read the first article then review the next two if you have time.
First review information we discussed in lecture today via this primer on immunity and herd immunity.
- Red and Orange team, Herd immunity: article 1, article 2, and article 3
- Yellow and Green team, vaccination: article 1, article 2, and article 3.
- Blue and Pink team, immunity through infection: article1, article 2, and article 3.
- Purple and Teal team, therapeutic antibodies: article 1, article 2, and article 3.
Round 2: Debrief Groups
Debrief groups will be assembled so people can share their expertise. Each group will contain at least one person from the four focus groups in round 1 and will be placed in a breakout together. Discuss the topics you were assigned in the context of moving quickly toward COVID-19 herd immunity. You'll take group notes via the google doc here. Please address the prompts and include your groups thoughts.
Round 3: Whole Group Discussion
Using our new insights and knowledge of herd immunity, we will discuss recent Massachusetts State policies that are designed to quickly reach herd immunity while providing vaccine rollout in an ethical way.
Policy 1: Effective Thursday, February 11th, an individual who accompanies a person age 75 or older to one of four mass vaccination sites to receive the vaccine will be eligible to receive the vaccine too, if they have an appointment booked. Both the companion and the individual age 75 or older must have an appointment for the same day and both individuals must be present. Only one companion is permitted. The companion must attest that they are accompanying the individual to the appointment.
See the state announcement here.
Policy 2:Effective Feb. 17, 2021 Massachusetts will be focusing its upcoming vaccine distribution efforts on its mass vaccination sites, such as Gillette Stadium and Fenway Park, and its partnerships with retail drugstores CVS and Walgreens. This means that venues such as MIT Medical are unlikely to receive vaccine in the coming weeks. See the announcement here.
Policy 3:Feb. 12th, Nearly every member of the Massachusetts congressional delegation is calling on Gov. Charlie Baker to set up a system to allow residents to pre-register and receive a notification for COVID-19 vaccine appointments.See news article here and copy of the letter here.
- Is the policy ethical?
- Is the policy a logical decision to get Massachusetts to herd immunity faster?
- Are there better approaches to this problem?
Additional reading:
- Antibody based COVID-19 treatments
- Cost of antibody based drugs
- CDC vaccine facts
- Ethical dilemmas of COVID-19 vaccination
- Debating duration of immunity
- Rocky road to herd immunity
- Choice to favor mass vaccination sites over hospitals
- Massachusetts $4.7m vaccine equity initiative
Reagents
- Chemically competent E. coli NEB5a (genotype: fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17)
- SOC medium: 2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose
- LB+Amp plates
- Luria-Bertani (LB) broth: 1% tryptone, 0.5% yeast extract, and 1% NaCl
- Plates prepared by adding 1.5% agar and 100 μg/mL ampicillin (Amp) to LB
- QIAprep Spin Miniprep Kit (from Qiagen)
- buffer P1
- buffer P2
- buffer N3
- buffer PB
- buffer PE
Reagents list
- scFv sequencing primers (Genewiz)
Next day: Analyze clone sequences and choose clone to characterize
Previous day: Harvest clone plasmids from library