Difference between revisions of "20.109(S17):Examine quantitative PCR results (Day9)"
MAXINE JONAS (Talk | contribs) (→Part 1: Practice statistical analysis) |
MAXINE JONAS (Talk | contribs) (→Part 2: Analyze quantitative PCR data) |
||
| Line 22: | Line 22: | ||
===Part 2: Analyze quantitative PCR data=== | ===Part 2: Analyze quantitative PCR data=== | ||
| − | As discussed in prelab, the qPCR experiment with your samples was not successful. This is likely due to degraded RNA, which is a very common issue given the prevalence, abundance, and stability of | + | As discussed in prelab, the qPCR experiment with your samples was not successful. This is likely due to degraded RNA, which is a very common issue given the prevalence, abundance, and stability of RNases. You will therefore use the data collected by the teaching faculty for this exercise and in your M2 Research Article. |
Before you can apply the statistical tools from Part 1 to your data, you must first normalize the p21 expression levels. To account for any unintended biases in RNA purification and / or cDNA preparation, it is important to normalize the expression of the transcript of interest to expression of a housekeeping or constitutive gene. Ideally, the gene to which the data of interest are normalized is not responsive to the treatment tested. In our experiment, we used GAPDH because it is not expected to be responsive to etoposide treatment. How might you confirm this assumption? | Before you can apply the statistical tools from Part 1 to your data, you must first normalize the p21 expression levels. To account for any unintended biases in RNA purification and / or cDNA preparation, it is important to normalize the expression of the transcript of interest to expression of a housekeeping or constitutive gene. Ideally, the gene to which the data of interest are normalized is not responsive to the treatment tested. In our experiment, we used GAPDH because it is not expected to be responsive to etoposide treatment. How might you confirm this assumption? | ||
Revision as of 13:21, 11 April 2017
Contents
Introduction
Today is the final laboratory session for Module 2! You have completed all of the bench work for your research; however, there is still data analysis to complete for the cell viability assay and the qPCR experiment. In addition to plotting and normalizing the data, you will complete statistical analysis to determine the significance of your results.
Statistics are mathematical tools used to analyze, interpret, and organize data. The specific tools that you will use are confidence intervals (CI) and the Student's t-test. To begin, review the following definitions:
- mean
- true mean
- standard deviation
Confidence intervals...error bars
Student's t-test...p-value
Protocols
Part 1: Practice statistical analysis
Review these data from an experiment where cells were exposed to increasing amounts of radiation. Your goal is to determine if a statistically significant amount of DNA damage was induced. For the purpose of this exercise, the values in the spreadsheet are in arbitrary units of 'DNA damage', where the higher numbers indicate more damage.
When interpreting the statistics, consider how you may use the information to convince someone that the DNA damage was significant. You may find this spreadsheet, originally created by Prof. Bevin Engelward and modified by the 20.109 staff, helpful for this exercise. At a minimum, you should post a bar plot of the data with 95% confidence intervals and indicate if there is a statistically significant difference (i.e. provide a p-value) between conditions in your Benchling notebook.
Part 2: Analyze quantitative PCR data
As discussed in prelab, the qPCR experiment with your samples was not successful. This is likely due to degraded RNA, which is a very common issue given the prevalence, abundance, and stability of RNases. You will therefore use the data collected by the teaching faculty for this exercise and in your M2 Research Article.
Before you can apply the statistical tools from Part 1 to your data, you must first normalize the p21 expression levels. To account for any unintended biases in RNA purification and / or cDNA preparation, it is important to normalize the expression of the transcript of interest to expression of a housekeeping or constitutive gene. Ideally, the gene to which the data of interest are normalized is not responsive to the treatment tested. In our experiment, we used GAPDH because it is not expected to be responsive to etoposide treatment. How might you confirm this assumption?
- Review the data in this spreadsheet.
- The DLD-1 and BRCA2- RNA isolated from untreated and etoposide treated cells was probed using both the p21 and GAPDH primers.
- Each reaction was completed in triplicate. Note: these are technical replicates.
- The data are represented as the 'threshold cycle' or amplification cycle at which SYBR Green fluorescent signal was detected (review M2D4 Introduction).
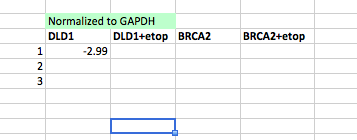
- Normalize p21 expression to GAPDH expression (ΔCT).
- Subtract the GAPDH CT value from the p21 CT value using the appropriate treatment conditions, according to the screenshot below.
- Exponentially transform each normalized value to the ΔCT expression.
- ΔCT expression = 2-ΔCT.
- Average the replicates for each treatment, then calculate the CI and p-value.
- With this information, graph your data with error bars and include information concerning any statistical significance.
- Are these results consistent with those from the RNA-seq data?
- Load the data: load("~/Desktop/RNA-seq data analysis/preprocessed_data.RData")library("DESeq2")
- Plot the reads for p21/CDKN1A: plotCounts(dds,"CDKN1A", intgroup="group")
Part 3: Analyze cell viability assay data
Review your cell viability results from M2D4 (posted to the Discussion tab of the M2 homepage). Use the statistical tools you learned in the above exercises to analyze the pooled class data for your M2 Research Article.
From these data, you can address two important experimental questions:
Next day: First day of M3
Previous day: Journal club II