20.109(S17):Complete cell survival assay and examine transcript levels in response to DNA damage (Day 4)
Contents
Introduction
Today you will be completing two experiments for module 2. The cell viability assay will provide information concerning the conditional lethality of compound 401 (NHEJ inhibitor) and olaparib (PARP inhibitor). In addition, you will use quantitative PCR to assess the levels of specific transcripts in response to DNA damage.
Cell viability assay
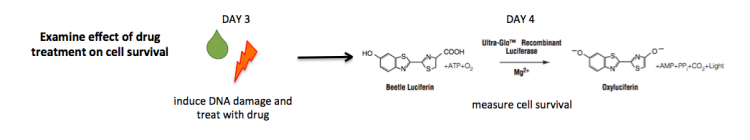
The CellTiter-Glo Luminescent Cell Viability Assay is a method for quantifying the number of viable cells based on measuring the amount of ATP present. ATP is a proxy for the presence of metabolically active (alive) cells. In this assay, the cells are lysed and ATP is released from the active cells. In a reaction catalyzed by a propriety luciferase enzyme, luciferin, ATP, and oxygen result in oxyluciferin, AMP, PPi, carbon dioxide, and light. The light product is then measured using a luminometer.
Quantitative PCR
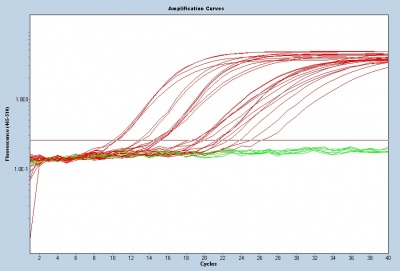
Quantitative polymerase chain reaction (qPCR) allows researchers to monitor the results of PCR as amplification is occurring (this technique is also referred to as real-time polymerase chain reaction or real-time PCR). During qPCR data are collected throughout the amplification process using a fluorescent dye. The fluorescent dye is highly specific for double-stranded DNA and when bound to DNA molecules the fluorescence intensity increases proportionately to the increase in double-stranded product. In contrast, the data for traditional PCR are simply observed as a band on a gel.
To assess gene transcript levels, you will examine the CT values from your qPCR assay. The CT values are displayed as an amplification curve following qPCR (these values are also given numerically). The initial cycles measure very little fluorescence due to low amounts of double-stranded DNA and are used to establish the inherent background fluorescence. As double-stranded product is produced, fluorescence is measured and the curve appears linear. This linear portion of the curve represents the exponential phase of PCR. Throughout the exponential phase, the curve should be smooth. Sharp points may be due to errors in reaction preparation or failures in the machine used to measure fluorescence. As mentioned previously, the first cycle in which the fluorescence measurement is above background is the CT. During the later cycles the curve shows minimal increases in fluorescence due the depletion of reagents.
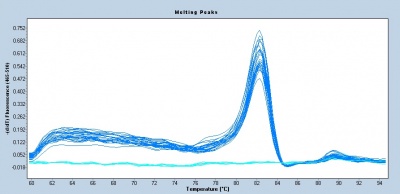
Following the qPCR amplification measurements, a melt curve is completed. Melt curves assess the dissociation of double-stranded DNA while the sample is heated. As the temperature is increased, double-stranded DNA ‘melts’ as the strands dissociate. As discussed above, the fluorescent dye used in qPCR associates with double-stranded DNA and fluorescence measurements will decrease as the temperature increases. In qPCR, the melt curve is used to confirm that a single amplification product was generated during the reaction. If additional products were present, the melt curve would presumably show additional peaks. Why might this be true? Can you think of a scenario where two different products would produce a single peak in a melt curve?
Protocols
Part 1: Complete cell viability assay
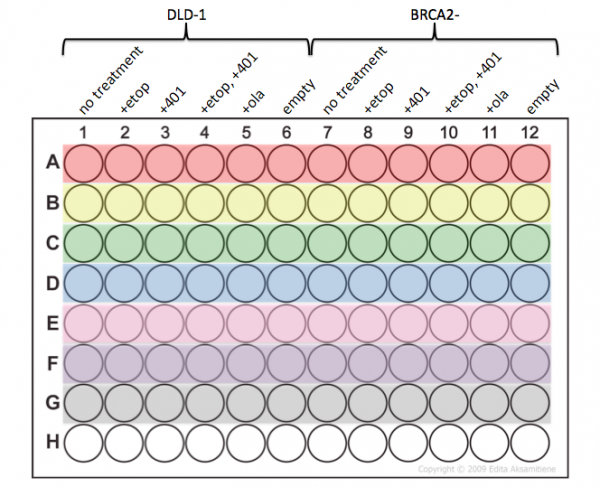
Today you will complete the second experiment for Mod 2 - the cell viability assay. In the previous class you used etoposide to induce DNA damage and added a NHEJ inhibitor. In addition, you treated your cells with a PARP inhibitor. By comparing the results of these treatments between the DLD-1 and BRCA2- cell lines you will more deeply examine the DNA repair pathways from lecture.
- Retrieve your plate from the 37 °C incubator.
- Aspirate the spent media from each well.
- Be careful not to cross-contaminate between the wells.
- Add 500 μL of fresh media into each well.
- You should also add media to the well that was empty as this will be your background control sample.
- Obtain an aliquot of the CellTiter Glo reagent from the front laboratory bench.
- Thoroughly mix the CellTiter Glo reagent, then add 500 μL into each well of your plate.
- After adding the reagent, pipet up and down two times to mix.
- Move your plate to the plate shaker at the front laboratory bench and shake for 2 min.
- Remove your plate from the plate shaker and incubate on your benchtop for 10 min.
- Transfer 100 μL from each well into a white 96-well plate.
- Be very careful that you add your samples to the appropriate wells according the plate map below.
The teaching faculty will take the plate to the BioMicro Center to measure the luminescence from your samples using the Varioskan plate reader.
Part 2: Design quantitative PCR primers
In Mod 1 you designed primers to amplify the gene that encodes FKBP12. As you may have guessed, quantitative PCR also requires primers for amplification. Because designing qPCR primers is more complicated (you need to carefully consider exons, product length, and primer dimers), you will use a free online program to identify possible primer pairs.
- Use the NCBI Primer-BLAST tool to design your qPCR primers.
- Go to the NCBI Gene Database.
- Enter your gene of interest (p21) in the search bar at the top of the screen.
- Review the list of the search results, and select one that is appropriate for our study (i.e. from what species should the gene be that you select?).
- Choose a gene sequence by clicking on the 'Name/Gene ID' link.
- Review the information provided on the page.
- What role does this gene product have in physiology?
- Why is this an interesting target for your research?
- In what tissues is expression of this gene highest? lowest?
- On which chromosome is it located?
- Scroll through the page to the NCBI Reference Sequences (RefSeq) section.
- Click on the link for the mRNA sequence (e.g. NM_000389.4).
- Record the mRNA sequence identification number in your laboratory notebook.
- Again, briefly review the information provided on the page.
- What is the size of the p21 gene?
- Is this sequence a variant? If so, how?
- Click on the 'Pick Primers' link under the Analyze this sequence header at the right side of the screen.
- Confirm that the correct mRNA sequence identification number is in the 'Enter accession, gi, or FASTA sequence' box under the PCR Template header at the top of the screen.
- Update the following settings:
- Under the Primer Parameters header, to the right of “PCR product size” enter a max number of 150.
- Under the Exon/intron selection header, select "Primer must span an exon-exon junction".
- Click the 'Get Primers' button at the bottom of the screen.
- Be patient! It may took up to 2 min for the program to identify possible primer pairs.
- Review the primer pairs identified by the program and consider the following guidelines:
- Primers should have a GC content of 50-60%.
- Primers ideally end in G or C.
- Primer melting temperatures should be similar and ~60 °C.
- Product should be ~100 bp.
- Select the primer pair that you think best meets the guidelines and record the sequences in your laboratory notebook.
For timing reasons, the teaching faculty previously ordered and tested qPCR primers that target p21.
- p21_F = CCA GCT GAG GTG TGA GCA G
- p21_R = GTT CTG ACA TGG CGC CTC C
You will use these primers for your reaction today, but should evaluate the sequences of these primers and compare them to those you selected. Did you choose the same primer pair? If not, what differences exist and which do you propose would be better at probing p21 mRNA sequences? Why?
Part 3: Setup quantitative PCR assay
In the previous laboratory session you purified RNA from your DLD-1 and BRCA2- cells and used the RNA to generate cDNA. Today you will use quantitative PCR to probe the transcription levels of genes of interest. Specifically, you will examine the response of DNA damage indicators to evaluate the etoposide treatment. In addition, you will ensure that exon 11 of BRCA2- is not expressed in the null mutant cell line... and that it is expressed in the parental cell line.
Part 3a: Clean cDNA
- Retrieve your cDNA samples from the front laboratory bench.
- Add 100 μL of Buffer PB to each cDNA preparation.
- Place four QIAquick columns into collection tubes and carefully label each according to your cDNA samples.
- Transfer the entire volume of cDNA + Buffer PB (~120 μL) from each tube into the appropriate QIAquick column.
- Centrifuge at maximum speed for 30 s, then discard the flow-through.
- Add 750 μL of Buffer PE to the QIAquick columns, centrifuge at maximum speed for 30 s, then discard the flow-through.
- Centrifuge the QIAquick columns at maximum speed for 1 min.
- This will remove any residual liquid from the columns.
- Move each QIAquick column into a clean, labeled 1.5 mL centrifuge tube.
- Be sure to remove the caps so they do not break off in the centrifuge.
- To elute the DNA, add 50 μL of dH2O pH = 8 to the center of the column and leave at your benchtop for 1 min.
- Centrifuge at maximum speed for 1 min.
- The liquid in the microcentrifuge tube is your cleaned cDNA.
- Alert the teaching faculty when you have finished and you will be escorted to the NanoDrop to measure the concentration of your cDNA samples.
Part 3b: Prepare cDNA for quantitative PCR assay
In addition to probing p21 transcript levels, you will also use primers specific for GAPDH. GAPDH (glyceraldehyde 3-phosphate dehydrogenase) is an enzyme that catalyzes a step of glycolysis, which serves to break down glucose for energy and carbon molecules. Because GAPDH expression should not be altered by etoposide treatment we will use its transcript levels to normalize the p21 expression levels between the DLD-1 and BRCA2- cell lines.
- Label 8 eppendorf tubes according to the designations below:
- A = DLD-1, GAPDH primers
- B = DLD-1 +etoposide, GAPDH primers
- C = BRCA2-, GAPDH primers
- D = BRCA2- +etoposide, GAPDH primers
- E = DLD-1, p21 primers
- F = DLD-1 +etoposide, p21 primers
- G = BRCA2-, p21 primers
- H = BRCA2- +etoposide, p21 primers
- Using the concentration measurements from Part 3a, calculate the volume of each sample that contains 20 ng of cDNA.
- If the volume is greater than 8 μL alert the teaching faculty.
- Prepare master mixes of your samples such that:
- Each reaction should contain 20 ng of cDNA, 2 μL of the appropriate diluted primer solution, and water for a total volume of 10 μL.
- The volume of water is calculated as 8 μL minus the volume calculated in Step #2 for each sample.
- For each sample/primer set combination, prepare enough master mix for 3.5 reactions.
- Give the tubes with your master mixes to the teaching faculty
Each master mix will be used to prepare three reactions (10 μL each), such that your samples are run in triplicate. This will allow you to complete statistics when you analyze the data on M2D9. To each 10 μL of sample the teaching faculty will add 10 μL of 2X SYBR Green reagent.
For your reference, the PCR cycling conditions are listed below:
| Stage | Cycles | Details |
|---|---|---|
| 1 | 1 | 95°C for 10 min |
| 2 | 40 | 95°C for 15 sec 60°C for 30 sec 72°C for 30 sec |
| 3 | 1 | 4°C hold |
Reagents
- growth medium (Life Technologies) for DLD-1 and BRCA2-/- cells
- RPMI 1640
- 10% fetal bovine serum (FBS)
- 100X antibiotic solution from Cellgro
- 10,000 U/mL penicillin
- 10,000 U/mL streptomycin
- CellTiter Glo cell viability assay kit (Promega)
- QIAquick PCR purification kit (Qiagen)
- primers
- GAPDH forward: GAA AGC CTG CCG GTG ACT AA
- GAPDH reverse: GCC CAA TAC GAC CAA ATC AGA G
- p21 forward: CCA GCT GAG GTG TGA GCA G
- p21 reverse: GTT CTG ACA TGG CGC CTC C
Next day: Journal club I
Previous day: Purify RNA for quantitative PCR assay and treat cells for survival assay