20.109(S07):Start-up genome engineering
Contents
Introduction
How many genetically-encoded creatures exist in every milliliter of sea water? 100? 1000? More? It turns out that bacteria are by far the best represented life form, numbering up to a million cells/ml. If each cell is assumed to harbor the DNA content of pedestrian E. coli MG1655, then that means 10^12th base pairs of DNA/ml. This thriving gene pool is even more remarkable in light of the fact that each ml contains approximately10^10th viruses that infect bacteria, aka bacteriophage or “phage” for short. As you might guess from that number, most of the earth’s bacteriophage are completely uncharacterized, though genome sequencing efforts are underway.
A few bacteriophage are exquisitely well characterized. Indeed, the study of phage laid much of the groundwork for our current understanding of genetics and molecular principles. These principles carry over to the biology of more complex cells (Jacques Monod famously said “What is true for Escherichia coli is true for the elephant” [Francois Jacob (1988. A statue within. Basic Books, New York, NY]). M13 is a member of the filamentous phage family. It has a long (~900 nm), narrow (~20 nm) protein coat that encases a small (~6.4 kb) single stranded DNA genome. The genome encodes 11 proteins, five of which are exposed on the phage’s protein coat and six of which are involved in phage maturation inside its E. coli host.
Phage Particles
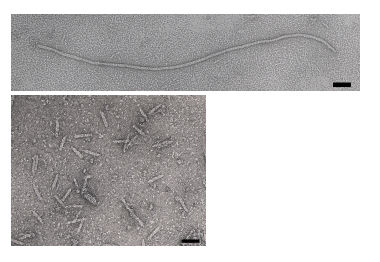
The phage coat is primarily assembled from a 50 amino acid protein called pVIII (or p8), which is sensibly enough encoded by geneVIII (or g8) in the phage genome. For a wild type M13 particle, it takes about ~2700 copies of p8 to make the ~900 nm long coat. There is lots of flexibility though and the number of p8 copies adjusts to accommodate the size of the single stranded genome it packages. When the phage genome was mutated to reduce its number of DNA bases (from 6.4 kb to 221 in the image below[[2]], image courtesy of Esther Bullitt, Boston University School of Medicine), then the p8 coat “shrink wraps’’ around the reduced genome, decreasing the number of p8 copies to less than 100. Electron micrographs of the resulting “microphage” and its wild type parent are shown below, where the black bar in each image is 50 nm long. And what about the upper limit to the length of the phage particle? Anecdotally, viable phage seems to top out at approximately twice the natural DNA content. However, deletion of a phage protein (p3) prevents full escape from the host E. coli, and phage that are 10-20X the normal length with several copies of the phage genome can be seen shedding from the E. coli host (look at the image on the coverpage to this module).
There are four other proteins on the phage surface, two of which have been extensively studied. At one end are five copies of the surface exposed pIX (p9) and a more buried companion protein, pVII (p7). These form the “blunt” end of the phage that’s seen in the micrographs and can be thought of as the eraser end of a pencil, with p8 forming the shaft. These proteins are some of the smallest known (only 33 and 32 amino acids), though some additional residues can be added to N-terminal portion of each and then presented on the “outside” of the phage coat (much more on this technique later). At the other end of the phage particle are five copies of the surface exposed pIII (p3) and it’s less exposed accessory protein, pVI (p6). These form the rounded tip of the phage and are the first proteins to interact with the E. coli host during infection and the last point of contact with the host as new phage bud from the bacterial surface.
Phage life-cycle
Protocols
- lab practical
- examine/troubleshoot M13K07 genomic map
- design oligos for epitope tagging (order?)
- digest bkb with Pst or Bam
DONE!
