Difference between revisions of "20.109(F22):M2D5"
Noreen Lyell (Talk | contribs) (→Introduction) |
Noreen Lyell (Talk | contribs) (→Introduction) |
||
| Line 6: | Line 6: | ||
==Introduction== | ==Introduction== | ||
| − | Interactions between low molecular weight ligands and proteins have been shown to increase the thermostability of proteins. This means that proteins bound to ligand are able to maintain tertiary structure, or resist denaturation, at higher temperatures than unbound proteins. Today we will use differential scanning fluorimetry (DSF) to examine the putative PfFKBP35 small molecule binders. | + | Interactions between low molecular weight ligands, or small molecules, and proteins have been shown to increase the thermostability of proteins. This means that proteins bound to ligand are able to maintain tertiary structure, or resist denaturation, at higher temperatures than unbound proteins. Today we will use differential scanning fluorimetry (DSF) to examine the putative PfFKBP35 small molecule binders. |
DSF is a method used to identify low molecular weight ligands that bind and stabilize a protein of interest. In this assay, protein denaturation is measured via a fluorescent dye that has an affinity for hydrophobic regions. When the protein is folded the hydrophobic pockets are inaccessible to the dye and the fluorescent signal is quenched by water in the solution. As the protein unfolds, the dye interacts with the hydrophobic regions and emits a fluorescent signal that can be detected. | DSF is a method used to identify low molecular weight ligands that bind and stabilize a protein of interest. In this assay, protein denaturation is measured via a fluorescent dye that has an affinity for hydrophobic regions. When the protein is folded the hydrophobic pockets are inaccessible to the dye and the fluorescent signal is quenched by water in the solution. As the protein unfolds, the dye interacts with the hydrophobic regions and emits a fluorescent signal that can be detected. | ||
Revision as of 18:01, 19 October 2022
Introduction
Interactions between low molecular weight ligands, or small molecules, and proteins have been shown to increase the thermostability of proteins. This means that proteins bound to ligand are able to maintain tertiary structure, or resist denaturation, at higher temperatures than unbound proteins. Today we will use differential scanning fluorimetry (DSF) to examine the putative PfFKBP35 small molecule binders.
DSF is a method used to identify low molecular weight ligands that bind and stabilize a protein of interest. In this assay, protein denaturation is measured via a fluorescent dye that has an affinity for hydrophobic regions. When the protein is folded the hydrophobic pockets are inaccessible to the dye and the fluorescent signal is quenched by water in the solution. As the protein unfolds, the dye interacts with the hydrophobic regions and emits a fluorescent signal that can be detected.
When a protein is bound to a ligand, the stability can be increased such that the temperature at which the protein denatures is increased. In the DSF assay, this is measured as a shift in the Tm, or melting temperature; which is defined as the temperature at which 50% of the protein is unfolded. This value represents the midpoint of the transition from structured (folded) to denatured (unfolded).
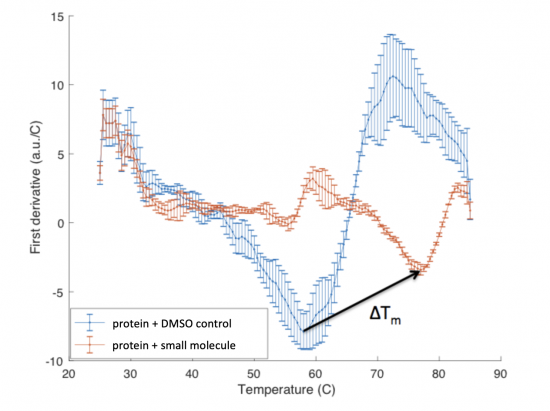
The ΔTm is the difference between the Tm of the unbound protein sample, or protein sample without added ligand, and the bound protein sample, protein sample with added ligand. If the tested ligand binds the protein of interest, the ΔTm can be observed as a shift in the plotted DSF data. For example, the data below show results of a pilot experiment completed in preparation for this module. In this graph the Tm of FKBP12 (blue curve) is ~50 °C. With the addition of rapamycin (red curve) the Tm is shifted to ~78 °C resulting in a ΔTm of ~20 degrees.
Protocols
Reagents list
- DSF dye (Thermo Fisher)
- ligands (Chembridge)
Next day: Complete data analysis of functional assay results