20.109(F20):M1D6
Contents
Introduction
Protocols
Part 1: Complete CometChip experiment
- Remove your CometChip from the 1X PBS and use a kimwipe to dry the GelBond side.
- Carefully move your CometChip to the gel electrophoresis station in the 4 °C cold room.
- Place your CometChip on the raised center region of an electrophoresis box.
- Double-sided tape was applied to the gel electrophoresis box. Be sure you lay your CometChip on the tape strips and lightly press down with a pipet tip to ensure it is secure.
- Add enough of the alkaline electrophoresis buffer to the gel electrophoresis box to cover your CometChip.
- Leave the CometChip in the alkaline electrophoresis buffer (aka unwinding buffer) for 45 minutes.
- To separate the damaged DNA into 'comets', electrophorese for 40 min (at 16 V, or 1 V/cm).
- It is important that the electrophoresis occur at 300 mA. To maintain the appropriate current, the volume of electrophoresis buffer may need to be adjusted. The teaching faculty will assist you in adding/removing electrophoresis buffer such that this value is reached.
- Carefully remove your CometChip from the electrophoresis box and place it in a dish.
- Obtain an aliquot of neutralization buffer from the front laboratory bench.
- Wash your CometChip by adding enough neutralization buffer to cover (~10-12 mL) and incubate for 5 min at room temperature.
- Repeat this step for a total of 3 washes.
- Add the SYBR gold DNA stain to your CometChip and carefully move it to the 4 °C cooler.
The teaching faculty will image your CometChip and provide the images to you in the next laboratory section.
Part 1: Analyze CometChip images
First, download your team's CometChip images from the Class data tab or obtain images from an instructor. Create (or copy and paste) the folder of your team images into the Documents\MATLAB\CometChip Analysis directory of the lab computer.
You will complete the following processes twice (stack images, optimize analysis parameters, measure tail lengths, export to Excel)--once for each CometChip. You should end up with one Excel file for each chip.
Stack images using ImageJ
- The script you will run will combine images from the same well into a stacked image and rename the files for the subsequent Matlab script to recognize them.
- Open ImageJ from Applications.
- Go to Plugins → Macros → Run...
- Select script "GenImageStacks_singleimage.txt" within the Documents\MATLAB\CometChip Analysis directory
- Choose the appropriate source directory that contains your image files.
- Find in Documents\MATLAB\CometChip Analysis directory
- Create a destination directory by selecting New Folder and naming the folder appropriately (eg. "180927M059_H2O2_stacked").
- Click Choose.
- ImageJ will create the stacked images in ~2 min.
- Please do not hit any additional keys until this process is completed.
- Confirm that the stacks are in the destination directory.
- Open the directory you just created, containing your stacked images.
- You should see one .tif image stack per well.
- Close ImageJ.
Optimize analysis parameters using MATLAB
- Open MATLAB from Applications.
- Be sure that "CometChip Analysis" is the current folder (double click on the "CometChip Analysis" folder from within the "Current Folder" window).
- In the command window, type in "guicometanalyzer" press enter to run the script.
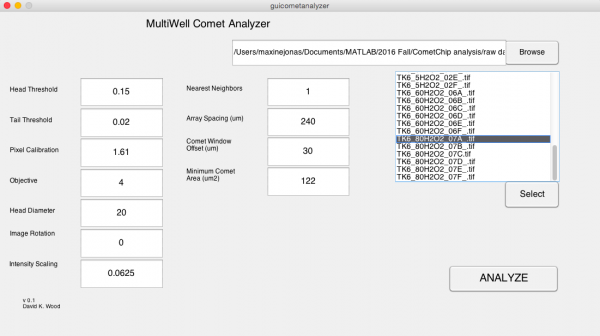
- In the MultiWell Comet Analyzer window:
- Check that the Pixel Calibration value is 1.61.
- Check that the Image Rotation value is 0 if your tails are to the right. Image Rotation should be 180 if your tails are going to the left.
- Check that the Array Spacing (μm) is 240.
- Check that the Head Diameter value is 20.
- Confirm the settings with the image below.
- Click Browse.
- Select the stacked image directory you created in the previous section.
- Click Open.
- From the image stack files loaded to the MultiWell Comet Analyzer window, choose ONE image that is expected to show DNA damage. You are just using this image to optimize the analysis parameters.
- The chosen file should be highlighted.
- Click Select.
- Click Analyze.
- Click "Yes" when the dialog box appears and asks if you want to run the program in "debug mode."
- Please do not hit any additional keys until this process is completed.
- In the MATLAB command window you will see the number of comets to be analyzed. The same number of figures should be generated.
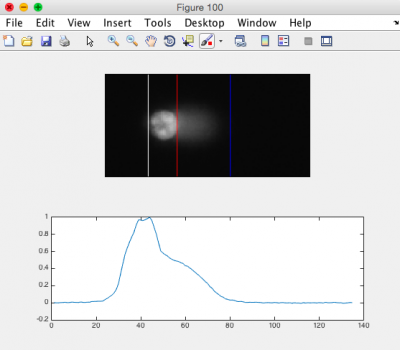
- Review the generated images.
- Ensure that the head of the comet is bracketed by a white line on the left, and a red line on the right. The tail should be bracketed by the red line and a blue line on the right. See the image to the right for an example.
- You may need to adjust the "Tail Threshold" value so that the tail is identified appropriately and/or the "Head Threshold" value so that the head is identified appropriately.
- Note: it is okay if some of the images do not appear correct according to the above criteria; however, the majority of your images should be correct.
- In the Command Window type "close all."
- If you need to adjust your parameters, repeat this process again until you find the optimal parameters for your analysis.
- Find the guicometanalzyer window.
- Adjust parameters as needed and repeat steps above by continuing to run in "debug mode."
- Type "close all" when done, but leave Matlab open.
Measure head and tail lengths using MATLAB
- Find the guicometanalyzer window
- If it has disappeared, run the script again by typing "guicometanalyzer" in the command window.
- In the MultiWell Comet Analyzer window, confirm the parameters as above.
- Click Browse.
- Select the stacked image directory you created with ImageJ.
- Click Open.
- Highlight all stack files in the MultiWell Comet Analyzer window.
- Click Select.
- Click Analyze.
- Click "No" when the dialog box appears and asks if you want to run the program in "debug mode."
- Confirm that there is a .txt file for each stack in your folder.
- Be sure to keep MATLAB open.
Compile results and export to Excel
- In the command window, type "comettoexcel" and press enter to run the script.
- When prompted, select the directory containing all the txt files that were output by guicometanalyzer and click "Open."
- The script will compile all your data, calculate the median values for each well, and export the data in an Excel file.
- Next, a dialog box will prompt you to save the Excel file.
- Please choose an appropriate directory and file name to save your data.
- Click "Save" and ensure your data has saved correctly.
Post your final Excel spreadsheets on the Class data tab.
Reagents list
- alkaline electrophoresis solution: 0.3 M NaOH, 1 mM Na2EDTA, pH 13.5 (from Sigma)
- neutralization buffer: 0.4 M Tris, pH 7.5 (from Sigma)
- SYBR gold DNA stain (from ThermoFisher Scientific)
Next day: Complete data analysis using statistical methods