20.109(F17):Develop experiment to optimize loading variables (Day2)
Contents
Introduction
Today you will load cells into the CometChip you prepared during the previous class to test cell loading conditions in an effort to optimize the assay developed by the Engelward Laboratory for our experimental purposes.
Cell loading refers to the process by which the cells are added to the CometChip. Briefly, you will add media containing cells over the agarose-based CometChip, and gravity will pull the cells down and 'load' them into the CometChip. The experimental details of this process will be the basis of the experiments you design and perform during this laboratory meeting.
Experimental design refers to the process by which the details of an experiment are organized to ensure that the data collected are appropriate and answer the correct question. In an experiment, a treatment is intentionally imposed on a sample such that the outcome(s) can be observed. You should consider these points when designing an experiment:
- Treatments should be administered in measurable levels. The level, or amount, of treatment must be conserved across samples to limit unintended variability in the results.
- Controls should be included. The controls, or untreated samples, are a baseline to which the treated samples are compared.
- Each experiment should have only one variable. If multiple variables (e.g. treatments, conditions, etc) are included in a single experiment, the results will be inconclusive because the outcome may be attributed to any of the variables within the experiment.
- Replicates should be included. The replicates test for technical error introduced by the researcher.
Designing a successful experiment requires time, effort, and practice. Today you will design an experiment to interrogate the best conditions for loading cells into your CometChip.
Protocols
Part 1: Design experiment to optimize CometChip loading
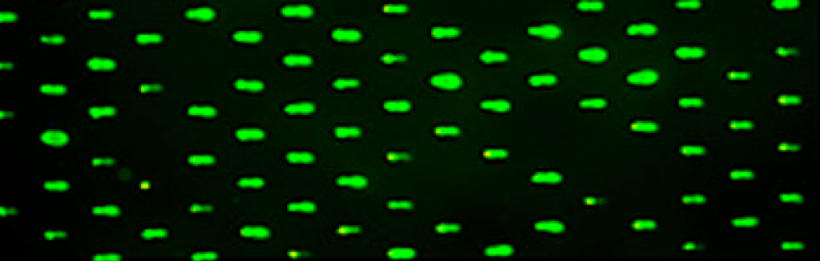
The experiment you design will test a variable associated with loading cells into the microwells of your CometChip. The variable in this experiment is cell number. Specifically, how many cells should be added to each well to ensure the majority of the microwells are loaded? In addition, how many cells should be loaded into each microwell?
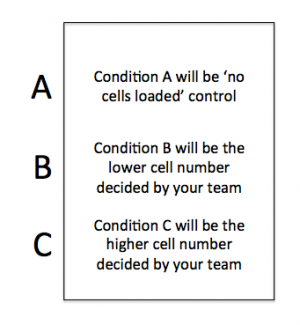
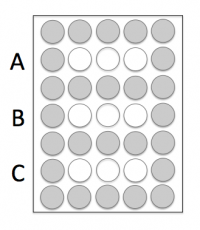
On your CometChip there is space for you to complete this experiment using three conditions. Your experiments will address the questions above and the conditions will provide data that will, hopefully, answer your research question.
Experiment: Determine the number of cells needed to completely load the microwells of your CometChip
- How many cells should be loaded into each microwell?
- Consider the amount of DNA that is carried by a single mammalian cell and the detection limit provided by the SYBR gold DNA stain that will be used in your experiments. Also, use the data you collected during the Orientation exercise to determine how many cells can fit into a single microwell based on the dimensions provided above.
- Be sure to include any calculations or thoughts in your laboratory notebook!
- When you know how many cells you want to load into each microwell, move on to the next question.
- How many cells should be added to each well such that the desired number of cells are loaded into the microwells?
- Consider the likelihood that every cell you add to the well will fall into a microwell. Perhaps calculate the surface area of the bottom of a well (of diameter 6.35 mm) and compare this to the size of the cells as you consider this question.
- Now that you have an idea as to the number of cells that are ideal for loading, consider the conditions you will use in your experiment.
- If you and your partner are in agreement on how many cells are ideal, you can load the ideal number of cells for Condition A and then either too few or too many for Condition B. Or you can load slightly more than the ideal number of cells for Condition A and slightly less for Condition B.
- If you and your partner are not in agreement, you can each load the number of cells you determine to be ideal as Condition A and Condition B.
- Again, all information concerning your experimental design choices should be recorded in your laboratory notebook!
Part 2: Prepare cells for CometChip loading
Part 3: Load CometChip
You will prepare the TK6 cells in the tissue culture hood and load them into the CometChip in the main laboratory. It is important that you consider the following details before entering the tissue culture room.
- The density of the TK6 cell suspension you will receive is 500,000 cells / mL.
- Calculate the volume of this suspension that you will need to load for Experiment 1. Note that this value will be different for Condition A and Condition B. If the calculated volumes are less than 50 μL or greater than 400 μL, consult the teaching faculty with how to move forward.
- You will also load cells into the wells for Experiment 2. For Experiment 2, you will load the same number of cells in all of the wells (remember from the introduction that each experiment should only contain a single variable). Select either Condition A or Condition B from Experiment 1 for loading here. If you and your partner are unable to agree on which Condition to use, consult the teaching faculty.
- The loading time will dictate when you add the cells to the wells of your CometChip.
- You will first load cells to the wells with the longest loading time, then add 100 μL 1x PBS to the wells you will use to test shorter loading times.
- Incubate your CometChip at 37 °C until the time difference between Condition A and Condition B has elapsed. For example, if Condition A is a 30 min incubation and Condition B is a 5 min incubation, load cells into the wells labeled for Condition A and 100 μL of 1x PBS into the wells labeled for Condition B. Move the CometChip to the 37 °C incubator and wait 25 min (30 min - 5 min = 25 min). Retrieve your CometChip from the incubator, aspirate the 1x PBS and add cells to the wells labeled for Condition B. Return the CometChip to the incubator for 5 min.
- In Experiment 2 you will add the same number of cells to all wells. As above, select the number of cells used for either Condition A or Condition B of Experiment 1 and/or consult the teaching faculty for advice.
Now that you have a plan, alert the teaching faculty. Because there are not enough biosafety cabinets for all the groups you may be asked to wait until another team has finished. Please be patient.
- Obtain the 15 mL conical of TK6 cells labeled with your group information.
- Note: the cells that you seeded last time were transferred to a 15 mL conical and the density was adjusted such that the suspension is 500,000 cells / mL.
- Retrieve your CometChip from the 4 °C cooler.
- You will also need to gather one glass plate, one 96-well bottomless plate, and four 1.5" binder clips from the front bench.
- Remove your CometChip from the 1x PBS and place it, gelbond side down, on the glass plate.
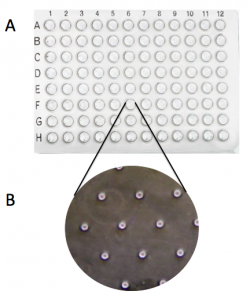
- Press the 96-well bottomless plate onto the CometChip so that the wells line up with your labeling as shown in the diagram on the right.
- Be sure to press the top of the 96-well bottomless plate onto the CometChip. If you are unsure which side is the top, please ask the teaching faculty.
- Do not move the 96-well bottomless plate while it is on the CometChip as you will damage the agarose and the microwells.
- Use the binder clips to secure the 96-well bottomless plate to the glass plate, thus creating a 'sandwich' with your CometChip in the center.
- Fasten the binder clips to the very edge of 96-well bottomless plate as shown in the image below.
- Add cells into the wells that will incubate for the longest loading time.
- Add 100 μL of 1x PBS to the empty wells.
- Incubate your CometChip at 37 °C for the appropriate length of time.
- Retrieve your CometChip.
- Remove the 1x PBS by aspirating off the liquid, gently, with a yellow pipet tip attached to the pasteur pipet.
- Be very careful not to damage the agarose!
- Press the pipet tip to the side of the well rather than poking down in the center of the well.
- Remove the pipet tip as soon as the 1x PBS is aspirated.
- Add cells into the appropriate wells and incubate at 37 °C for the length of time specified by your experimental condition.
- After the cells are loaded, complete a wash step to remove excess cells that are not within the microwells of your CometChip. Read all the bullets below before proceeding!
- Carefully remove the binder clips and the 96-well bottomless plate.
- Alert the teaching faculty at this point! The wash step can be very temperamental and it is best to see a demonstration!
- With the CometChip on the glass plate, 'waterfall' ~5 mL of 1x PBS over the wells, which are now imprinted onto the agarose.
- To waterfall the 1x PBS, hold the glass plate at a 45° angle over the dish that you used to store your CometChip.
- Pipet up ~5 mL of 1x PBS.
- Press the pipet tip onto the glass plate above your CometChip.
- As you expel the 1x PBS, quickly move the pipet tip from left-to-right.
- The 1x PBS should pass over the top of the CometChip and fall into the dish.
- Use a yellow tip attached to the pasteur pipet to aspirate the excess liquid from your CometChip wells.
- Lightly touch the tip to the bottom of each imprinted well on the CometChip and immediately lift the tip from the agarose.
- Read through Steps #13-16 before continuing with the procedure.
- Retrieve one tube of molten 0.3% low melting point (LMP) agarose from the 42 °C waterbath.
- You will need to work quickly from this point as the LMP agarose will solidify as it cools.
- 1% penicillin/streptomycin antibiotics were added to the LMP agarose to prevent bacterial contamination.
- Using the P1000, pipet up 1000 μL of molten agarose from the tube.
- Hold the pipet tip over the top left well of your CometChip and as you expel the agarose move the pipet tip from left to right. Complete this process for every row of your CometChip.
- The goal is to lightly cover the wells that contain cells, which will 'trap' the cells into the microwells.
- Leave your CometChip undisturbed in the biosafety cabinet for 15 min.
- Repeat Steps #13-16 for a second layer of 0.3% LMP agarose.
- If the LMP agar 'fell' off the CometChip in any areas during this process, it is important to 'fill in' those portions of the CometChip. Please alert the teaching faculty if you experience any difficulties!
- Cover your CometChips and carefully move them to the 4 °C cooler for 15 min.
- USE MICROSCOPE TO VIEW CELLS / WELL
- CELL LYSIS ON
Part 4: Practice statistics analysis
Reagents list
CometChip:
- agar, normal melting point (Invitrogen)
- phosphate buffered saline (VWR)
- GelBond film (Lonza)
- 1 well dish (VWR)
- agar, low melting point (Invitrogen)
- bottomless 96-well plates (VWR)
- 10% formalin (VWR)
- Vybrant® DyeCycle™ Green Stain (Invitrogen)
Cell culture:
- RPMI 1640 (Invitrogen)
- with 10% fetal bovine serum (Atlanta Biologicals)
- 100X antibiotic solution (Invitrogen)
- 10,000 U/mL Penicillin
- 10,000 U/mL Streptomycin
- Trypan Blue
- Incubator maintains 37°C, 5% CO2 and 95% relative humidity
Next day: Evaluate cell loading results and prepare biochemical experiment