20.109(S19):Module 2
Contents
Module 2
Lecturers: Ernest Fraenkel and Noreen Lyell
Instructors: Noreen Lyell and Leslie McClain
TAs: Catherine Henry and Michaela Gold
Lab manager: Hsinhwa Lee
Overview
Homologous recombination (HR) and non-homologous end joining (NHEJ) are two pathways of DNA repair by which double-strand breaks are corrected in mammalian cells. In specific types of cancer, targeting a DNA repair pathway is an effective treatment option as a paradox exists in cancer. Whole genome sequencing has revealed that the cells of many tumor types have mutations in genes necessary for DNA repair. These mutations are responsible for cells becoming cancerous, but are also detrimental because, just like normal cells, cancer cells must divide to survive. Thus, a cancer cell will develop an ‘addiction’ to a DNA repair pathway – specifically, a pathway different from the one with the mutation that caused the cell to generate a tumor. Recent cancer therapies seek to exploit this addiction by targeting the intact pathway used by the tumor cells to repair DNA damage due to intrinsic breaks that occur during replication. In addition, the effectiveness of DNA damage induced by chemotherapy treatment may be enhanced by also disrupting the functional repair pathways of tumor cells.
In previous semesters students have studied this phenotype by treating cancer cells that are deficient in HR with drugs that target NHEJ enzymes. Surprisingly, in their data the treatment appeared to improve survival in the cancer cells!
In this module, you will analyze transcriptome data and design a cell viability experiment to examine the effect of drug treatment and the BRCA2-/- genetic mutation on cancer cells given this result. To this end, you will address two primary research questions:
- How does treatment with a chemotherapy drug alter gene expression in cancer cells?
- How does a BRCA2 mutation alter gene expression in cancer cells?
An additional goal of Module 2 is for you to synthesize data from different experiments into a single story. More often that not, researchers reach conclusions using results gathered from more than one experimental approach. Multiple layers of support can bolster a conclusion as they eliminate biases that may exist in a particular method of analysis. Furthermore, for complex questions it may be necessary to include data from several approaches to provide a sufficient conclusion.
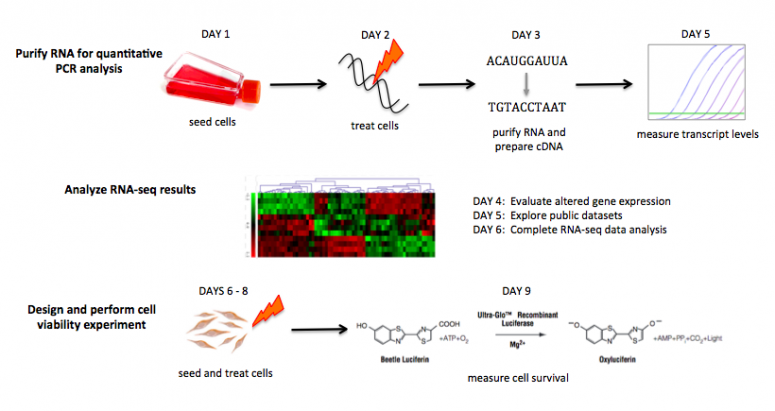
Lab links: day by day
M2D1: Practice tissue culture techniques and prepare cells for RNA purification
M2D2: Etoposide treat cells for RNA purification
M2D3: Purify RNA and practice RNA-seq data analysis methods
M2D4: Analyze RNA-seq data and prepare for quantitative PCR experiment
M2D5: Perform quantitative PCR experiment and explore additional RNA-seq datasets
M2D6: Design cell viability experiment and complete RNA-seq data analysis
M2D7: Journal club I presentations
M2D8: Journal club II presentations
M2D9: Complete data analysis
Assignments
Journal club presentation
Research article
References
- A syngeneic variance library for functional annotation of human variation: application to BRCA2. Cancer Research. 68:5023-5030.
- Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood Journal. 117:6074-6082.