20.109(F18):Module 1
Contents
Module 1
Lecturer: Bevin Engelward
Instructors: Noreen Lyell, Leslie McClain and Josephine Bagnall
TAs: Corban Swain and Jai Padmakumar
Lab manager: Hsinhwa Lee
Overview
Cancer is a disease of the genome. Cancer is caused by accumulated mutations in genes that lead to phenotypic advantages in the progression from normal to metastatic cancer. While we know a lot about the kinds of changes that cancer cells undergo, we do not yet fully understand where mutations come from in the first place. What is clear is that changes to the DNA structure (e.g., DNA damage) can cause carcinogenic mutations. Given that DNA damage causes mutations that drive cancer progression, it is important to have effective tools for measuring DNA damage. In this module, you will learn about two different approaches for quantifying DNA damage. The first method relies on physical changes to the DNA structure that impact its ability to migrate when electrophoresed. This is a direct measure of DNA damage. The second method relies on antibody recognition of changes to proteins that occur as a result of cell signaling that is triggered by DNA damage. This is an indirect measure of DNA damage. Given the potentially deadly consequences of DNA damage, it is a good thing that our cells have robust ways to repair their DNA. Being able to measure DNA damage means that we can study DNA repair, which turns out to be a very important variable when it comes to why some people get cancer, and others do not.
In addition to learning many fundamental biological and engineering concepts, you will of course learn many laboratory techniques. In particular, you will learn about electrophoresis, mammalian cell culture, quantitative image analysis techniques, basic statistics, enzyme kinetics, molecular pathway analysis, impact of genetic factors, epigenetic analysis, antibody labeling and much more.
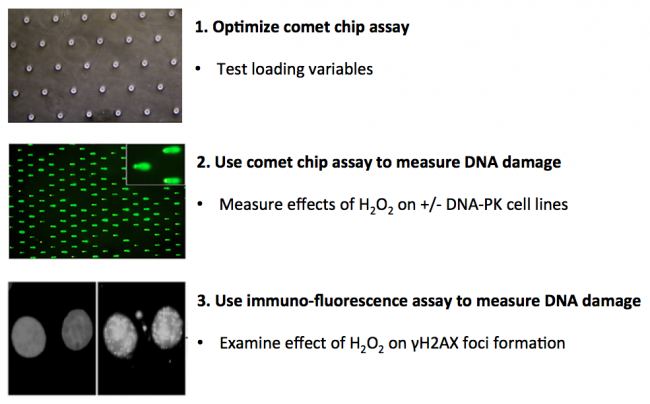
In terms of the specific experiments that you will do, in this module you will measure genomic instability using two techniques: a microwell array (CometChip) and a sub-nuclear foci assay (γH2AX immunofluorescence). Your first task is to critically think through the development of the CometChip assay and determine which conditions provide the best results for loading mammalian cells into the device. To this end, you will consider variables that affect cell loading into the microwells of the CometChip. The data you collect will be used to determine the conditions for subsequent assays. Next, you will use the CometChip assay to measure DNA damage in response to a deficiency in repair and a chemical treatment. Specifically, you will study the role of DNAPKcs in DNA repair and the effects of oxidative stress on DNA damage via H2O2 treatment. Then, you will use the γH2AX to visualize DNA damage in individual cells and compare the results to the CometChip data.
Lab links: day by day
M1D1: Practice tissue culture and prepare microwell array
M1D2: Design cell loading optimization experiment and research cell lines
M1D3: Prepare and treat cells for genomic instability experiment
M1D4: Complete genomic instability experiment and load cells for sub-nuclear foci assay
M1D5: Analyze genomic instability experiment data and treat cells for sub-nuclear foci assay
M1D6: Complete sub-nuclear foci assay
M1D7: Practice statistical analysis methods and complete data analysis
Assignments
Data summary
Mini-presentation
References
CometChip: A high-throughput 96-well platform for measuring DNA damage in microarrayed human cells. Journal of Visualized Experiments. (2014) 92: 1-11.
- A video of the procedure is linked here.
CometChip: Single-cell microarray for high-throughput detection of DNA damage. Methods in Cell Biology. (2012) 112: 247-268.