Difference between revisions of "Protocols for cell culture"
From Course Wiki
MAXINE JONAS (Talk | contribs) |
MAXINE JONAS (Talk | contribs) |
||
| Line 4: | Line 4: | ||
{{Template:20.309}} | {{Template:20.309}} | ||
| + | ==Preparing Solutions== | ||
| − | == | + | ====DMEM reconstitution==== |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | For 1 L of DMEM++ | |
| − | + | * In the hood, combine | |
| − | + | ** 1 L DMEM | |
| − | + | ** 100 mL FBS | |
| + | ** 10 mL Penicillin/Streptavidin | ||
| + | * Filter into sterile container. | ||
| + | * Label as DMEM++ and store at 4°C. | ||
| − | + | ====3.7% Formaldehyde==== | |
| + | * One 16% ampoule | ||
| + | * 3.3 mL 1x PBS | ||
| + | Store in dark at room temperature. | ||
| − | == | + | ====0.1% Triton X-100==== |
| + | * 0.05 g Triton X-100 | ||
| + | * 50 mL PBS | ||
| + | Store at room temperature. | ||
| − | + | ====1% BSA==== | |
| − | + | * 0.5 g BSA | |
| − | + | * 50 mL PBS | |
| − | + | Store at 4°C. | |
| − | + | ||
| − | + | ||
| − | + | ====10 μM cytochalasin D==== | |
| + | * 50 μL 2mM cytoD | ||
| + | * 10 mL PBS | ||
| + | Store at -20°C. | ||
| − | + | ====Alexa Fluor dye solution==== | |
| − | + | *7 μL dye in methanol | |
| − | + | *200 μL PBS | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | == | + | ====Microsphere cleaning==== |
| − | + | ||
| − | + | # For 10 mL of bead solution at a concentration of 5e5 beads/mL: Pipet 1 μL of bead solution and 99 μL of molecular-grade water into centrifuge tube. Vortex. | |
| − | + | # Centrifuge the microspheres at 1300 g for 10 minutes to clear the supernatant. | |
| + | # Remove and discard supernatant. Resuspend the microspheres in 100 μL of water and vortex. | ||
| + | # Centrifuge at 1300 g for 10 minutes. | ||
| + | # Pipet out supernatant and bring into tissue culture hood. Resuspend microspheres in 1 mL of DMEM++ to get concentration of 5e6 beads/mL. (Check in hemocytometer.) Allocate 9 mL of DMEM++ into Falcon tube. Vortex the beads and transfer into the same Falcon tube. Vortex the 10 mL solution. Beads should be at a concentration of 5e5 beads/mL. | ||
| + | # Before pipetting onto cells: vortex the solution, sonicate for 10 minutes to break up aggregates, and warm in hot water bath. | ||
| + | |||
| + | |||
| + | ==Splitting cells== | ||
| + | |||
| + | # Turn on 37° water bath, open tissue culture hood, turn on light and vent, and spray counter with 70% ethanol. Periodically spray gloves with ethanol throughout the procedure. | ||
| + | # In the hood, pipette 125 mL of DMEM++ into a separate sterile container. Warm the DMEM++, PBS, and Trypsin containers in the bath. | ||
| + | # After warming up for 20 minutes, take DMEM++, PBS, and Trypsin out of bath. Dry them, wipe with ethanol, and bring into hood. | ||
| + | # Add 25 mL of DMEM++ into the new T75 flask. | ||
| + | # Fetch flask of cells from incubator and make sure they look ok under the inverted microscope. Spray with ethanol and bring into hood. | ||
| + | # Load a glass pipette into narrow end of vacuum tube. Attach wide end of tube to the vacuum line. Use the glass pipette to drain the medium from a corner of the old flask. With the 5 mL pipette, rinse old flask with 4 mL of PBS. Drain with vacuum. | ||
| + | # Pipette 2 mL of Trypsin into old flask and close the cap. Set timer for 4 minutes and put in incubator. Use the microscope to verify cell detachment. | ||
| + | # Spray flask with ethanol and bring into hood. Pipette 25 mL of DMEM++ into flask of trypsinized cells. Make sure to spread the medium and wash the cells from the walls by pipetting up and down. Using the same pipette, transfer less than 1 mL of cells into an eppendorf tube. Take eppendorf out of hood. | ||
| + | # Transfer 90 μL of cells and 10 μL of Trypan blue dye into new eppendorf. Load each chamber of the hemacytometer with ~15 μL of the cell-dye mixture. | ||
| + | # There should be 9 large squares in the field of view. Count the number of cells in the 4 large corner squares and the large center square. Repeat with second chamber and sum the two counts. Multiply by 1000 to calculate the number of cells/mL. | ||
| + | # Calculate the volume required for the desired number of reseeded cells, and pipette that volume into new T75 flask. To plate the MatTeks, first add more DMEM++ into the flask of cells such that there are approximately 5x10<math>^4</math> in 0.5 mL. Add 1.5 mL of medium into each MatTek dish. Pipette 0.5 mL of cells into each MatTek. Close the caps, label, and place in incubator. | ||
| + | # Cleanup: | ||
| + | * Vacuum remainder of cells. Drain 50 mL Falcon tube of bleach to wash the system. Disconnect vacuum tubes. | ||
| + | * Put extra DMEM++ and PBS in the fridge. Trysin goes in the -20°C freezer. | ||
| + | * Wipe the counter and close the hood. Clean hemacytometer. Toss eppendorf, old flasks, and gloves in the biowaste container. | ||
| + | |||
| + | |||
| + | ==Fixing and labeling cells== | ||
| + | |||
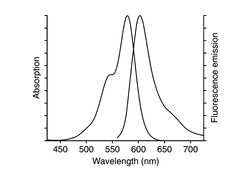
| + | [[File: Alexa568.jpg|thumb|250px|Excitation and emission spectra for Alexa Fluor 568.]] | ||
| + | Prepare a dish of fixed fibroblast cells (NIH 3T3), whose actin will be labeled with phalloidin-Alexa Fluor 568. The labeling protocol is as follows: | ||
| + | * Start with cells cultured in dishes containing DMEM++, at approximately 60% confluency. This is about the optimum percentage of cell population. If cells are too crowded, they will not stretch properly and show their beautiful actin filaments. Note also that these cells remain alive until the addition of formaldehyde, therefore requiring that any buffer/media added be pre-warmed. | ||
| + | |||
| + | # Pre-warm 3.7% formaldehyde solution, DMEM and PBS in a 37°C water bath. Keep the solution wrapped in foil to protect from light. | ||
| + | # Retrieve an aliquot of cytochalasin D from the freezer and warm in the water bath to 37°C. | ||
| + | # After you get your dishes from the TA or Instructor, first remove the medium with a pipette and wash each dish twice with 2 mL of pre-warmed phosphate buffered saline (PBS) at pH 7.4 (also in the water batch). Pipet up and down into the dish gently to avoid washing away cells. | ||
| + | # Carefully pipet 400 μL of 3.7% formaldehyde solution onto the central glass region of the dish and leave it for 10 minutes at room temperature. This "fixes" the cells, i.e. cross-links the intracellular proteins and freezes the cell structure. | ||
| + | # Wash the cells 3X with 2 mL PBS (note that this PBS solution no longer needs to be pre-warmed as the cells are dead). | ||
| + | # Extract each dish with 2 mL 0.1% Triton X-100 (a type of soap solution in PBS) for 3-5 minutes. (Extraction refers to partially dissolving the plasma membrane of the cell.) | ||
| + | # Wash the cells 3X with PBS. | ||
| + | # Incubate the fixed cells with 2 mL 1% BSA in PBS for 20 minutes. (BSA blocks the nonspecific binding sites.) | ||
| + | # Wash cells 2X with PBS. | ||
| + | # To each cell dish, add 200 μL of Alexa Fluor 568 phalloidin solution (specific binder to F-actin) pre-mixed in methanol and PBS. Carefully pipet this just onto the center of the dish, cover with aluminum foil, and incubate for 45 minutes at room temperature. | ||
| + | # Wash 3X with PBS. | ||
| + | # You can now store the sample at +4°C (regular refrigerator) in PBS for a few days, wrapped in parafilm and foil. It can also be stored in mounting medium for up to 1 year. | ||
| − | + | By a very similar procedure, you should also prepare cells treated with cytochalasin D (CytoD): Before adding the formaldehyde, add 1 mL of the pre-warmed 10 μM CytoD solution to the cell culture dish and incubate at 37°C for 20 minutes. Afterwards, wash 2X with DMEM++. | |
| − | + | ||
| − | |||
==References== | ==References== | ||
Revision as of 15:01, 13 August 2013
Preparing Solutions
DMEM reconstitution
For 1 L of DMEM++
- In the hood, combine
- 1 L DMEM
- 100 mL FBS
- 10 mL Penicillin/Streptavidin
- Filter into sterile container.
- Label as DMEM++ and store at 4°C.
3.7% Formaldehyde
- One 16% ampoule
- 3.3 mL 1x PBS
Store in dark at room temperature.
0.1% Triton X-100
- 0.05 g Triton X-100
- 50 mL PBS
Store at room temperature.
1% BSA
- 0.5 g BSA
- 50 mL PBS
Store at 4°C.
10 μM cytochalasin D
- 50 μL 2mM cytoD
- 10 mL PBS
Store at -20°C.
Alexa Fluor dye solution
- 7 μL dye in methanol
- 200 μL PBS
Microsphere cleaning
- For 10 mL of bead solution at a concentration of 5e5 beads/mL: Pipet 1 μL of bead solution and 99 μL of molecular-grade water into centrifuge tube. Vortex.
- Centrifuge the microspheres at 1300 g for 10 minutes to clear the supernatant.
- Remove and discard supernatant. Resuspend the microspheres in 100 μL of water and vortex.
- Centrifuge at 1300 g for 10 minutes.
- Pipet out supernatant and bring into tissue culture hood. Resuspend microspheres in 1 mL of DMEM++ to get concentration of 5e6 beads/mL. (Check in hemocytometer.) Allocate 9 mL of DMEM++ into Falcon tube. Vortex the beads and transfer into the same Falcon tube. Vortex the 10 mL solution. Beads should be at a concentration of 5e5 beads/mL.
- Before pipetting onto cells: vortex the solution, sonicate for 10 minutes to break up aggregates, and warm in hot water bath.
Splitting cells
- Turn on 37° water bath, open tissue culture hood, turn on light and vent, and spray counter with 70% ethanol. Periodically spray gloves with ethanol throughout the procedure.
- In the hood, pipette 125 mL of DMEM++ into a separate sterile container. Warm the DMEM++, PBS, and Trypsin containers in the bath.
- After warming up for 20 minutes, take DMEM++, PBS, and Trypsin out of bath. Dry them, wipe with ethanol, and bring into hood.
- Add 25 mL of DMEM++ into the new T75 flask.
- Fetch flask of cells from incubator and make sure they look ok under the inverted microscope. Spray with ethanol and bring into hood.
- Load a glass pipette into narrow end of vacuum tube. Attach wide end of tube to the vacuum line. Use the glass pipette to drain the medium from a corner of the old flask. With the 5 mL pipette, rinse old flask with 4 mL of PBS. Drain with vacuum.
- Pipette 2 mL of Trypsin into old flask and close the cap. Set timer for 4 minutes and put in incubator. Use the microscope to verify cell detachment.
- Spray flask with ethanol and bring into hood. Pipette 25 mL of DMEM++ into flask of trypsinized cells. Make sure to spread the medium and wash the cells from the walls by pipetting up and down. Using the same pipette, transfer less than 1 mL of cells into an eppendorf tube. Take eppendorf out of hood.
- Transfer 90 μL of cells and 10 μL of Trypan blue dye into new eppendorf. Load each chamber of the hemacytometer with ~15 μL of the cell-dye mixture.
- There should be 9 large squares in the field of view. Count the number of cells in the 4 large corner squares and the large center square. Repeat with second chamber and sum the two counts. Multiply by 1000 to calculate the number of cells/mL.
- Calculate the volume required for the desired number of reseeded cells, and pipette that volume into new T75 flask. To plate the MatTeks, first add more DMEM++ into the flask of cells such that there are approximately 5x10$ ^4 $ in 0.5 mL. Add 1.5 mL of medium into each MatTek dish. Pipette 0.5 mL of cells into each MatTek. Close the caps, label, and place in incubator.
- Cleanup:
- Vacuum remainder of cells. Drain 50 mL Falcon tube of bleach to wash the system. Disconnect vacuum tubes.
- Put extra DMEM++ and PBS in the fridge. Trysin goes in the -20°C freezer.
- Wipe the counter and close the hood. Clean hemacytometer. Toss eppendorf, old flasks, and gloves in the biowaste container.
Fixing and labeling cells
Prepare a dish of fixed fibroblast cells (NIH 3T3), whose actin will be labeled with phalloidin-Alexa Fluor 568. The labeling protocol is as follows:
- Start with cells cultured in dishes containing DMEM++, at approximately 60% confluency. This is about the optimum percentage of cell population. If cells are too crowded, they will not stretch properly and show their beautiful actin filaments. Note also that these cells remain alive until the addition of formaldehyde, therefore requiring that any buffer/media added be pre-warmed.
- Pre-warm 3.7% formaldehyde solution, DMEM and PBS in a 37°C water bath. Keep the solution wrapped in foil to protect from light.
- Retrieve an aliquot of cytochalasin D from the freezer and warm in the water bath to 37°C.
- After you get your dishes from the TA or Instructor, first remove the medium with a pipette and wash each dish twice with 2 mL of pre-warmed phosphate buffered saline (PBS) at pH 7.4 (also in the water batch). Pipet up and down into the dish gently to avoid washing away cells.
- Carefully pipet 400 μL of 3.7% formaldehyde solution onto the central glass region of the dish and leave it for 10 minutes at room temperature. This "fixes" the cells, i.e. cross-links the intracellular proteins and freezes the cell structure.
- Wash the cells 3X with 2 mL PBS (note that this PBS solution no longer needs to be pre-warmed as the cells are dead).
- Extract each dish with 2 mL 0.1% Triton X-100 (a type of soap solution in PBS) for 3-5 minutes. (Extraction refers to partially dissolving the plasma membrane of the cell.)
- Wash the cells 3X with PBS.
- Incubate the fixed cells with 2 mL 1% BSA in PBS for 20 minutes. (BSA blocks the nonspecific binding sites.)
- Wash cells 2X with PBS.
- To each cell dish, add 200 μL of Alexa Fluor 568 phalloidin solution (specific binder to F-actin) pre-mixed in methanol and PBS. Carefully pipet this just onto the center of the dish, cover with aluminum foil, and incubate for 45 minutes at room temperature.
- Wash 3X with PBS.
- You can now store the sample at +4°C (regular refrigerator) in PBS for a few days, wrapped in parafilm and foil. It can also be stored in mounting medium for up to 1 year.
By a very similar procedure, you should also prepare cells treated with cytochalasin D (CytoD): Before adding the formaldehyde, add 1 mL of the pre-warmed 10 μM CytoD solution to the cell culture dish and incubate at 37°C for 20 minutes. Afterwards, wash 2X with DMEM++.