Difference between revisions of "Assignment 9, Part 1: Analyze two-color yeast images"
Juliesutton (Talk | contribs) (→Analyze a frame) |
MAXINE JONAS (Talk | contribs) m (→Yeast image analysis code) |
||
| (32 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | == | + | [[Category:20.309]] |
| + | [[Category:Electronics]] | ||
| + | {{Template:20.309}} | ||
| + | __NOTOC__ | ||
| + | ==Written questions== | ||
| + | {{Template:Assignment Turn In|message = | ||
| − | + | Read the paper and answer the written questions in [[Assignment 9 Overview: Analyzing yeast images]]. | |
| − | The sample is similar to the one in Mettetal et al.: we are using ''S. | + | }} |
| + | |||
| + | ==Yeast image analysis code== | ||
| + | |||
| + | Start by downloading Assignment9TestData.mat. This is an example of the data you will be collecting in Assignment 10 with an 8 minute valve oscillation period. The movie was collected using the same 20.309 microscope that you built in Assignment 8. During an oscillation experiment, the microscope first recorded an image with blue illumination followed by one with green illumination. It had a 40x objective and a 125 mm tube lens - remember that results in a 25x magnification. | ||
| + | |||
| + | The sample is similar to the one in Mettetal ''et al.'': we are using ''S. cerevisiae'' with the protein Hog1 fused to GFP (which is excited by blue light) and an mRNA binding protein (we'll call MCP) fused to tagRFP (which is excited by green light) to locate the nucleus. Our goal is to calculate the average correlation between Hog1 and the nucleus as a function of the high/low valve state. | ||
In MATLAB, display the contents of the structure in the data file: | In MATLAB, display the contents of the structure in the data file: | ||
| Line 19: | Line 30: | ||
The data structure contains all the important information from the experiment: | The data structure contains all the important information from the experiment: | ||
| − | * A two-color movie, where the 3d dimension is the color (1 = Blue, 2 = Green), and the fourth is the frame number. Notice that <tt>twoColorYeastTest.Movie</tt> has already converted to a double, but it is not yet normalized by 4095. | + | * A two-color movie, where the 3d dimension is the color (1 = Blue excitation = GFP, 2 = Green excitation = RFP), and the fourth is the frame number. Notice that <tt>twoColorYeastTest.Movie</tt> has already converted to a double, but it is not yet normalized by 4095. |
| − | * The time stamp of each frame | + | * The time stamp of each frame, recorded in seconds from the start of the experiment. Column 1 contains the timestamps of the GFP images, column 2 is for the RFP images. |
* The oscillation valve period, in seconds. | * The oscillation valve period, in seconds. | ||
| − | * The blue and green | + | * The blue and green illumination camera settings in the format <tt>[gain, exposure]</tt>, where <tt>exposure</tt> is in microseconds. |
* The valve state (0 for low salt, 1 for high salt) at the time of each image. If you need to know the valve state more precisely, it can be calculated with the following conditional statement: <math> sin(2 \pi t/T) \ge 0 </math>, where ''T'' is the valve oscillation period, and ''t'' is the time since the start of the experiment (this is how the state is determined in the software). | * The valve state (0 for low salt, 1 for high salt) at the time of each image. If you need to know the valve state more precisely, it can be calculated with the following conditional statement: <math> sin(2 \pi t/T) \ge 0 </math>, where ''T'' is the valve oscillation period, and ''t'' is the time since the start of the experiment (this is how the state is determined in the software). | ||
{{Template:Assignment Turn In|message = Choose a single frame of the movie. Make a four panel figure and display: the blue image, the green image (both with a scale bar), and their corresponding image histograms. Include an appropriate title for each panel. }} | {{Template:Assignment Turn In|message = Choose a single frame of the movie. Make a four panel figure and display: the blue image, the green image (both with a scale bar), and their corresponding image histograms. Include an appropriate title for each panel. }} | ||
| − | == | + | ==Estimating background levels== |
| − | + | [[File:BlueImageHistogram.png|thumb|right]] | |
| − | [[ | + | |
| − | + | It's tricky to get good images of fluorescent proteins expressed by living yeast cells. Not only do we need to use very high exposure times (on the order of 5 s) to capture a dim signal, but we want to image cells over long times (minutes to hours). These factors work against us to produce high background levels caused by autofluorescence of the surrounding medium, camera noise (especially dark noise), and potentially light leakage from the room. Any drift in the background level over time can obscure our results. Additionally, the background level may not be uniform over the entire image. For each frame of the movie, we will subtract off a background image (taken with no cells present) which has been scaled appropriately to account for changing levels over time. | |
| + | In the example data provided, you should find a background image taken with no yeast present - only background coming from the medium autofluorescence and other background sources. To determine how much we need to scale the image, we will compare the background image intensity to an empty region of our cell images. | ||
| + | |||
| + | Use this function to locate an empty region of the first frame in your movie: | ||
<pre> | <pre> | ||
| − | function | + | function BackgroundRectangle = LocateBackgroundRegion( InitialYeastImage ) |
| − | + | ||
| − | + | figure | |
| − | + | imshow( InitialYeastImage(:,:,1) ) | |
| − | + | title('Drag a rectangle around a region of the image containing only background') | |
| − | + | BackgroundRectangle = getrect( gcf ); | |
| − | + | close | |
| − | + | ||
| + | end | ||
| + | </pre> | ||
| + | Then for each frame, you can calculate the scale factor needed for your background image: | ||
| + | <pre> | ||
| + | function backgroundScaleFactors = FindBackgroundScaleFactor( InitialImage, BackgroundImage, BackgroundRectangle ) | ||
| + | backgroundScaleFactors = nan(1,2); | ||
| + | % Scale background | ||
| + | for j = 1:2 | ||
| + | backgroundImageCropped = imcrop(BackgroundImage(:,:,j),BackgroundRectangle); | ||
| + | initialImageCropped = imcrop(InitialImage(:,:,j),BackgroundRectangle); | ||
| + | backgroundScaleFactors(j) = mean(initialImageCropped(:))/ mean(backgroundImageCropped(:)); | ||
| + | end | ||
| + | |||
end | end | ||
</pre> | </pre> | ||
| − | + | Test the code by finding the background scale factors for the first frame of the GFP and RFP images. Try using several different rectangular regions and notice how much the output varies. | |
| + | {{Template:Assignment Turn In|message = Display an example of a background-subtracted and contrast-stretched image for a GFP and RFP image.}} | ||
| − | == | + | ==Analyze a frame== |
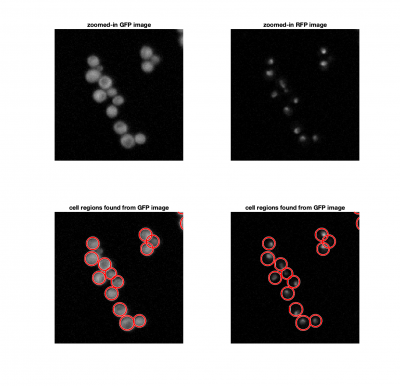
| − | + | In each frame of our movie, we need to locate and identify individual cells, and find the correlation coefficient between the cell image and the nuclei image. If the Hog1 signal is localized in the nucleus, the correlation should be close to 1. | |
| − | + | [[Image:FindingCellsAndNucleiExample.png|center|thumb|400px]] | |
| − | + | Start with the following basic layout of the function: | |
| − | + | ||
| − | + | ||
<pre> | <pre> | ||
| − | function [ | + | function [AverageNucleusHog1Correlation, StandardErrorNucleusHog1Correlation] = AnalyzeFrame( TwoColorFrame, BackgroundImage, BackgroundRectangle ) |
| − | + | ||
| − | + | % 1. estimate the background level | |
| − | + | backgroundScaleFactors = FindBackgroundScaleFactor(TwoColorFrame,BackgroundImage, BackgroundRectangle); | |
| − | + | ||
| − | + | cellFrame = TwoColorFrame(:,:,1) - backgroundScaleFactors(1)*BackgroundImage(:,:,1); | |
| − | + | nucleiFrame = TwoColorFrame(:,:,2) - backgroundScaleFactors(2)*BackgroundImage(:,:,2); | |
| − | + | ||
| − | + | % 2. identify individual yeast cells in the GFP image | |
| − | + | % 3. calculate the correlation coefficient between the GFP intensities an RFP intensities within each cell region | |
| − | + | % 4. output the mean correlation coefficient for all cells in the frame | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
end | end | ||
</pre> | </pre> | ||
| − | + | As you work through this part of the Assignment, fill out the <tt>AnalyzeFrame</tt> function to incorporate each of the outlined steps. | |
| − | ===Finding | + | ===Finding cells=== |
| − | We | + | We could consider using a threshold to identify cells in an image, similar to how we found fluorescent particles in Assignments 4 and 5. The problem here, is that thresholding doesn't allow us separate a single cells from a clump of cells. Since we know that yeast cells are roughly spherical, we can use <tt>imfindcircles</tt> to locate them. <tt>imfindcircles</tt> is an implemenattion of the [https://www.mathworks.com/help/images/ref/imfindcircles.html| Hough transform]. |
<pre> | <pre> | ||
| − | + | [cellCenters, cellRadii] = imfindcircles(cellFrame,[Rmin Rmax],'ObjectPolarity','bright','Sensitivity',0.95); | |
| − | + | ||
</pre> | </pre> | ||
| − | + | Here, <tt>Rmin</tt> and <tt>Rmax</tt> are the smallest and largest expected 'radii'. How can you estimate <tt>Rmin</tt> and <tt>Rmax</tt>? | |
| − | + | The function <tt>viscircles(cellCenters,cellRadii)</tt> may be useful to visualize your results. | |
| − | + | === Loop through each cell and calculate the correlation coefficient between GFP and RFP === | |
| − | + | Next we want to calculate the response of the Hog1 protein. In the paper, they calculated the response as the ratio of GFP intensities inside and outside the nucleus: <math> R_{Hog1} = \frac{<GFP_{nucleus}>}{<GFP_{cell}>}</math>. We'll use an alternative way to find the response: by finding the correlation coefficient between the GFP and RFP signals within each cell. | |
| + | Finish filling out <tt>AnalyzeFrame</tt> to loop through each circle found in the frame and extract the corresponding pixel intensities from your GFP and RFP images. Then, calculate the correlation coefficients between these two intensity vectors (<tt>corr</tt> in MATLAB). The following helper function may be useful. | ||
<pre> | <pre> | ||
| − | + | function Im = createMaskFromCircles(centers,radii,ImageSize) | |
| + | Im = zeros(ImageSize); | ||
| + | [X, Y] = meshgrid(1:ImageSize(2), 1:1:ImageSize(1)); | ||
| + | |||
| + | for ii = 1:length(radii) | ||
| + | isBright = (X-centers(ii,1)).^2+(Y-centers(ii,2)).^2 <= radii(ii,1)^2; | ||
| + | isAlreadyFound = (Im == 1); | ||
| + | Im = Im + (isBright & ~isAlreadyFound); | ||
| + | end | ||
| + | Im( Im > 1 ) = 1; | ||
| + | end | ||
</pre> | </pre> | ||
| − | + | Once you've collected each individual correlation coefficient, find the mean and standard error, and output these from the function. | |
| + | |||
| + | == Analyze the movie== | ||
| + | |||
| + | Once you've set up your analysis for a single frame, you'll now want to loop through the movie and collect the responses as a function of time. | ||
| + | |||
| + | {{Template:Assignment Turn In|message = | ||
| + | |||
| + | # Analyze each frame of the movie and plot the average cellular response as a function of time. | ||
| + | #* In a single figure, plot the data in one subplot, and the pinch valve state (1 for high salt, 0 for low salt) in a second subplot. | ||
| + | # Write a function to fit the response to a sinusoid and extract the predicted amplitude and phase shift of the response signal. Plot the best fit function on the same plot as your data. | ||
| + | # Report the resulting best fit parameters. }} | ||
| + | |||
| + | |||
| + | {{Template:Assignment Turn In|message = Turn in all your MATLAB code in pdf format. No need to include functions that you used but did not modify.}} | ||
| + | |||
| + | {{Template:Assignment Turn In|message = If you have not already done so,[[Assignment 8, Part 3: add flow control and test your device| complete questions 4 and 5 from Assignment 8]]. If you were not able to collect good step response data, either use a section of your longest oscillation movie data, or borrow some data from another group (and don't forget to give them credit!).}} | ||
| − | + | {{Template:Assignment 9 Analyzing yeast images navigation}} | |
| + | {{Template:20.309 bottom}} | ||
Latest revision as of 16:08, 12 November 2019
Written questions
| |
Read the paper and answer the written questions in Assignment 9 Overview: Analyzing yeast images. |
Yeast image analysis code
Start by downloading Assignment9TestData.mat. This is an example of the data you will be collecting in Assignment 10 with an 8 minute valve oscillation period. The movie was collected using the same 20.309 microscope that you built in Assignment 8. During an oscillation experiment, the microscope first recorded an image with blue illumination followed by one with green illumination. It had a 40x objective and a 125 mm tube lens - remember that results in a 25x magnification.
The sample is similar to the one in Mettetal et al.: we are using S. cerevisiae with the protein Hog1 fused to GFP (which is excited by blue light) and an mRNA binding protein (we'll call MCP) fused to tagRFP (which is excited by green light) to locate the nucleus. Our goal is to calculate the average correlation between Hog1 and the nucleus as a function of the high/low valve state.
In MATLAB, display the contents of the structure in the data file:
twoColorYeastTest =
struct with fields:
Movie: [544×728×2×16 double]
Time: [16×2 double]
ValveOscillationPeriod: 480
BlueCameraGainAndExposure: [5 5000000]
GreenCameraGainAndExposure: [15 5000000]
ValveState: [16×1 logical]
The data structure contains all the important information from the experiment:
- A two-color movie, where the 3d dimension is the color (1 = Blue excitation = GFP, 2 = Green excitation = RFP), and the fourth is the frame number. Notice that twoColorYeastTest.Movie has already converted to a double, but it is not yet normalized by 4095.
- The time stamp of each frame, recorded in seconds from the start of the experiment. Column 1 contains the timestamps of the GFP images, column 2 is for the RFP images.
- The oscillation valve period, in seconds.
- The blue and green illumination camera settings in the format [gain, exposure], where exposure is in microseconds.
- The valve state (0 for low salt, 1 for high salt) at the time of each image. If you need to know the valve state more precisely, it can be calculated with the following conditional statement: $ sin(2 \pi t/T) \ge 0 $, where T is the valve oscillation period, and t is the time since the start of the experiment (this is how the state is determined in the software).
Estimating background levels
It's tricky to get good images of fluorescent proteins expressed by living yeast cells. Not only do we need to use very high exposure times (on the order of 5 s) to capture a dim signal, but we want to image cells over long times (minutes to hours). These factors work against us to produce high background levels caused by autofluorescence of the surrounding medium, camera noise (especially dark noise), and potentially light leakage from the room. Any drift in the background level over time can obscure our results. Additionally, the background level may not be uniform over the entire image. For each frame of the movie, we will subtract off a background image (taken with no cells present) which has been scaled appropriately to account for changing levels over time.
In the example data provided, you should find a background image taken with no yeast present - only background coming from the medium autofluorescence and other background sources. To determine how much we need to scale the image, we will compare the background image intensity to an empty region of our cell images.
Use this function to locate an empty region of the first frame in your movie:
function BackgroundRectangle = LocateBackgroundRegion( InitialYeastImage )
figure
imshow( InitialYeastImage(:,:,1) )
title('Drag a rectangle around a region of the image containing only background')
BackgroundRectangle = getrect( gcf );
close
end
Then for each frame, you can calculate the scale factor needed for your background image:
function backgroundScaleFactors = FindBackgroundScaleFactor( InitialImage, BackgroundImage, BackgroundRectangle )
backgroundScaleFactors = nan(1,2);
% Scale background
for j = 1:2
backgroundImageCropped = imcrop(BackgroundImage(:,:,j),BackgroundRectangle);
initialImageCropped = imcrop(InitialImage(:,:,j),BackgroundRectangle);
backgroundScaleFactors(j) = mean(initialImageCropped(:))/ mean(backgroundImageCropped(:));
end
end
Test the code by finding the background scale factors for the first frame of the GFP and RFP images. Try using several different rectangular regions and notice how much the output varies.
| |
Display an example of a background-subtracted and contrast-stretched image for a GFP and RFP image. |
Analyze a frame
In each frame of our movie, we need to locate and identify individual cells, and find the correlation coefficient between the cell image and the nuclei image. If the Hog1 signal is localized in the nucleus, the correlation should be close to 1.
Start with the following basic layout of the function:
function [AverageNucleusHog1Correlation, StandardErrorNucleusHog1Correlation] = AnalyzeFrame( TwoColorFrame, BackgroundImage, BackgroundRectangle )
% 1. estimate the background level
backgroundScaleFactors = FindBackgroundScaleFactor(TwoColorFrame,BackgroundImage, BackgroundRectangle);
cellFrame = TwoColorFrame(:,:,1) - backgroundScaleFactors(1)*BackgroundImage(:,:,1);
nucleiFrame = TwoColorFrame(:,:,2) - backgroundScaleFactors(2)*BackgroundImage(:,:,2);
% 2. identify individual yeast cells in the GFP image
% 3. calculate the correlation coefficient between the GFP intensities an RFP intensities within each cell region
% 4. output the mean correlation coefficient for all cells in the frame
end
As you work through this part of the Assignment, fill out the AnalyzeFrame function to incorporate each of the outlined steps.
Finding cells
We could consider using a threshold to identify cells in an image, similar to how we found fluorescent particles in Assignments 4 and 5. The problem here, is that thresholding doesn't allow us separate a single cells from a clump of cells. Since we know that yeast cells are roughly spherical, we can use imfindcircles to locate them. imfindcircles is an implemenattion of the Hough transform.
[cellCenters, cellRadii] = imfindcircles(cellFrame,[Rmin Rmax],'ObjectPolarity','bright','Sensitivity',0.95);
Here, Rmin and Rmax are the smallest and largest expected 'radii'. How can you estimate Rmin and Rmax?
The function viscircles(cellCenters,cellRadii) may be useful to visualize your results.
Loop through each cell and calculate the correlation coefficient between GFP and RFP
Next we want to calculate the response of the Hog1 protein. In the paper, they calculated the response as the ratio of GFP intensities inside and outside the nucleus: $ R_{Hog1} = \frac{<GFP_{nucleus}>}{<GFP_{cell}>} $. We'll use an alternative way to find the response: by finding the correlation coefficient between the GFP and RFP signals within each cell.
Finish filling out AnalyzeFrame to loop through each circle found in the frame and extract the corresponding pixel intensities from your GFP and RFP images. Then, calculate the correlation coefficients between these two intensity vectors (corr in MATLAB). The following helper function may be useful.
function Im = createMaskFromCircles(centers,radii,ImageSize)
Im = zeros(ImageSize);
[X, Y] = meshgrid(1:ImageSize(2), 1:1:ImageSize(1));
for ii = 1:length(radii)
isBright = (X-centers(ii,1)).^2+(Y-centers(ii,2)).^2 <= radii(ii,1)^2;
isAlreadyFound = (Im == 1);
Im = Im + (isBright & ~isAlreadyFound);
end
Im( Im > 1 ) = 1;
end
Once you've collected each individual correlation coefficient, find the mean and standard error, and output these from the function.
Analyze the movie
Once you've set up your analysis for a single frame, you'll now want to loop through the movie and collect the responses as a function of time.
| |
Turn in all your MATLAB code in pdf format. No need to include functions that you used but did not modify. |
| |
If you have not already done so, complete questions 4 and 5 from Assignment 8. If you were not able to collect good step response data, either use a section of your longest oscillation movie data, or borrow some data from another group (and don't forget to give them credit!). |
- Overview
- Part 1: Analyze a two-color yeast movie frame
Back to 20.309 Main Page