Difference between revisions of "20.109(S24):M2D1"
Becky Meyer (Talk | contribs) (→Protocols) |
Becky Meyer (Talk | contribs) (→Part 5: Design primers for site-directed mutagenesis) |
||
| Line 50: | Line 50: | ||
*What amino acid | *What amino acid | ||
| − | ===Part 5: Design primers for site-directed mutagenesis=== | + | ===Part 5: Design primers for site-directed mutagenesis=== |
| + | INTRO-- SDM to insert sequence of interest | ||
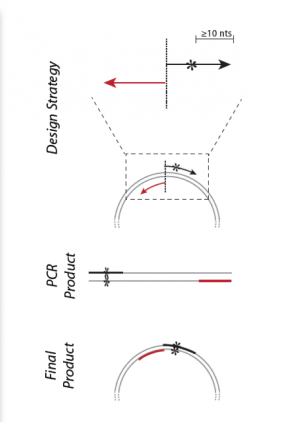
| − | + | [[Image:Sp16 M1D2 primer design schematic.png|thumb|right|300px| '''Schematic for inserting sequences in plasmids using SDM technique.''' Image modified from Q5 Site-Directed Mutagenesis Kit Manual published by NEB.]]To insert a new DNA sequence into an expression plasmid, synthetic primers can be used in a technique referred to site-direction mutagenesis (see figure on the right). Primer design for site-directed mutagenesis, or SDM, is quite straightforward: the forward primer (considered a mutagenic primer because it is changing the DNA) introduces new bases into the coding strand. As the plasmid is amplified during PCR, a population of plasmids will be created which incorporate the new bases. | |
| − | + | ||
| − | [[Image:Sp16 M1D2 primer design schematic.png|thumb|right|300px| '''Schematic for | + | |
Primers used in SDM must meet several design criteria to ensure specificity and efficiency. Consider the following design guidelines for mutagenesis primers: | Primers used in SDM must meet several design criteria to ensure specificity and efficiency. Consider the following design guidelines for mutagenesis primers: | ||
| − | *Desired mutation | + | *Desired mutation must be present in the forward primer. |
*Forward and reverse primers should 'face' away from the mutation and be 'back-to-back' when annealed to the template. | *Forward and reverse primers should 'face' away from the mutation and be 'back-to-back' when annealed to the template. | ||
*Primers should be 25-45 bp long. | *Primers should be 25-45 bp long. | ||
| Line 67: | Line 66: | ||
**where N is primer length and the two percentages should be integers | **where N is primer length and the two percentages should be integers | ||
| − | + | Luckily, online tools are available to assist with SDM primer design. Today you will use NEBaseChanger (provided by NEB) to design your primers. | |
| + | #Go to the [http://nebasechanger.neb.com/ NEBaseChanger] site and click 'Please enter a new sequence to begin.' | ||
| + | ***Then what? | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
*Include a screen capture of the information provided in the Result and Required Primers sections. | *Include a screen capture of the information provided in the Result and Required Primers sections. | ||
Revision as of 21:42, 23 February 2024
Contents
- 1 Introduction
- 2 Protocols
- 2.1 Part 1: Review metabolic engineering approach to create M17 yeast
- 2.2 Part 2: Review yeast cell surface display
- 2.3 Part 3: Examine data for M17 yeast cell surface amino acids
- 2.4 Part 4: Choose a peptide sequence and determine a DNA sequence to encode it
- 2.5 Part 5: Design primers for site-directed mutagenesis
- 3 Navigation links
Introduction
Heavy metals are typically thought of as metal elements with a relatively high density. Many of these metals are considered precious (gold, silver, platinum) or have commercial value (lead, copper, nickel). Other metals can play an essential health role in small quantities (zinc, iron, manganese). However many of these metals are toxic, even when present in parts per billion. Heavy metals can be released into the environment through a number of activities such as mining, industrial production of consumer products, or disposal of electronic waste.
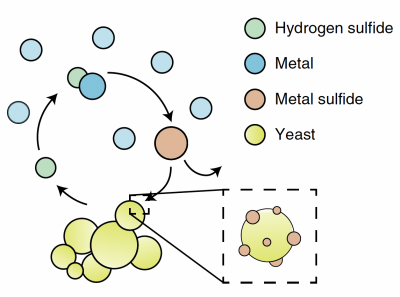
In this module, you will focus on protein engineering of a cell surface display peptide in Saccharomyces cerevisiae, also known as baker's yeast. We are using this YSD (yeast cell surface display) model to capture the heavy metal cadmium in a recyclable form. This type of protein engineering is an early step in optimizing a "bioremediation" model system. Bioremediation is an approach to cleaning up environmental pollutants by repurposing a known biological process. Typically, bioremediation utilizes microbial and fungal organisms to remove environmental contaminants as these single cell organisms can be fast growing and genetically tractable (i.e. relatively simple to genetically manipulate).
Today you examine what is known about our current S.cerevisiae bioremediation model system. You will explore the genetic metabolic engineering used to induce the yeast to produce hydrogen sulfide, examine the components of yeast cell surface display, and examine previously expressed peptides. You will use this information to rationally design a peptide to express on the yeast cell surface in order to capture Cadmium particles in the most uniform distribution possible. The rest of the module will be devoted to expressing your chosen peptides in our yeast model system, and testing these peptides for their suitability in bioremediation.
Protocols
Part 1: Review metabolic engineering approach to create M17 yeast
In this module,
In your laboratory notebook, complete the following:
- How
Part 2: Review yeast cell surface display
In the
In your laboratory notebook, complete the following:
- Based
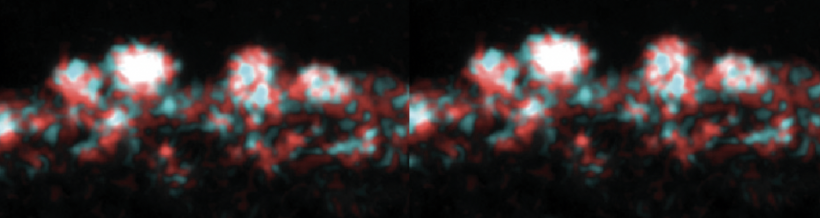
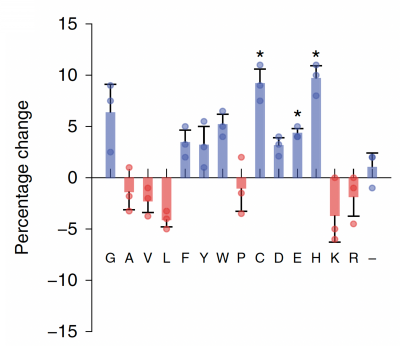
Part 3: Examine data for M17 yeast cell surface amino acids
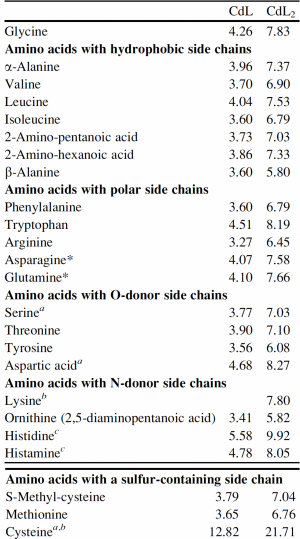
Now that you have noted relevant information regarding the Fet4 transporter, it's time to turn your attention to our metal of interest, cadmium. While there is limited information on the specific mechanism of transporter-mediated cadmium uptake into cells, previous literature has indicated that amino acid residues form complexes of differing stability with cadmium. Using the table to the right, examine the reported affinities of cadmium for different amino acid residues.In your laboratory notebook, complete the following:
- Based on the information above, which amino acids seem like likely candidates to capture precipitating cadmium?
In your laboratory notebook, complete the following:
- Which
Part 4: Choose a peptide sequence and determine a DNA sequence to encode it
Using the information you have gathered above, you can now determine what peptide you would like express in order to capture precipitating cadmium. ADD WAY MORE
In your laboratory notebook, complete the following:
- What amino acid
Part 5: Design primers for site-directed mutagenesis
INTRO-- SDM to insert sequence of interest
Primers used in SDM must meet several design criteria to ensure specificity and efficiency. Consider the following design guidelines for mutagenesis primers:
- Desired mutation must be present in the forward primer.
- Forward and reverse primers should 'face' away from the mutation and be 'back-to-back' when annealed to the template.
- Primers should be 25-45 bp long.
- G/C content of > 40% is desired.
- Both primers should terminate in at least one G or C base.
- Melting temperature should exceed 78°C, according to:
- Tm = 81.5 + 0.41 (%GC) – 675/N - %mismatch
- where N is primer length and the two percentages should be integers
Luckily, online tools are available to assist with SDM primer design. Today you will use NEBaseChanger (provided by NEB) to design your primers.
- Go to the NEBaseChanger site and click 'Please enter a new sequence to begin.'
- Then what?
In your laboratory notebook, complete the following:
- Include a screen capture of the information provided in the Result and Required Primers sections.
- Use the guidelines provided above to examine the mutagenesis primers designed by NEBaseChanger. Do the primers meet the design criteria?
Copy your forward and reverse primer sequence and upload them to the Class Data page on the wiki before you leave.
- These primers must be ordered tonight to arrive in time for your next experiment.
Next day: Perform site-directed mutagenesis