20.109(S21):M1D4
Introduction:
Today you will submit plasmids for sequencing analysis to determine the sequence differences between these clones and our characterized lysozyme binding scFv clone. These newly isolated plasmids encode for unique single-chain antibody fragment (scFv) clones that may show improved binding to our antigen, lysozyme.
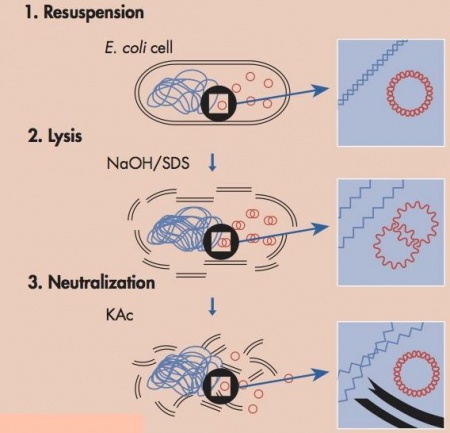
To harvest plasmid DNA from the yeast cells we will be using a kit that is based off the common E. coli plasmid purification method called alkaline lysis. For yeast there needs to be one additional step to help break down the cell wall, but overall goal of each prep is the same--to separate the plasmid DNA from the chromosomal DNA and cellular debris, allowing the plasmid DNA to be studied further. The key difference between plasmid DNA and chromosomal DNA is size and this difference is what is used to separate the two components.In the plasmid DNA purification protocol, the media is removed from the cells by centrifugation. The cells are resuspended in an solution that contains Tris to buffer the cells and EDTA to bind divalent cations in the lipid bilayer, thereby weakening the cell envelope. An alkaline (basic) lysis buffer is added that contains sodium hydroxide and the detergent sodium dodecyl sulfate (SDS). The base denatures the cell’s DNA, both chromosomal and plasmid, while the detergent dissolves the cellular proteins and lipids. The pH of the solution is returned to neutral by adding a mixture of acetic acid and potassium acetate. At neutral pH the SDS precipitates from solution, carrying with it the dissolved proteins and lipids. The DNA strands renature at neutral pH. The chromosomal DNA, which is much longer than the plasmid DNA, renatures as a tangle that gets trapped in the SDS precipitate. The smaller plasmid DNA renatures normally and stays in solution, effectively separating plasmid DNA from the chromosomal DNA and the proteins and lipids of the cell. At this point the solution is spun at a high speed and soluble fraction, including the plasmid, is kept for further purification and the insoluble fraction, the macromolecules and chromosomal DNA is pelleted and thrown away.
Protocols
Reagents list
- scFv sequencing primers (Genewiz)
Next day: [[]]