Difference between revisions of "20.109(F22):M1D4"
Noreen Lyell (Talk | contribs) (→Protocols) |
(→Introduction) |
||
| (23 intermediate revisions by 2 users not shown) | |||
| Line 5: | Line 5: | ||
==Introduction== | ==Introduction== | ||
| − | + | Today you will analyze data from your first experiment in 20.109! When collecting data from an experiment, it is important to be a neutral observer. This will prevent you from introducing bias into the results. Bias refers to any treatment that differs between experimental groups that does NOT relate to the experimental question. To maximize reliability of data, scientists are trained to limit experimental bias as much as possible. To give you an idea of some of the many ways bias can be introduced into an experiment, consider the following scenarios: | |
| − | + | *An experimenter uses the last remaining volume of buffer on their control samples. They make a new, fresh batch of buffer for their experimental samples. | |
| + | *An experimenter always loads in their control samples and experimental samples in the same order into a 96 well plate when using an automated sampling machine. | ||
| + | *An experimenter treats all of their control samples, then all of their experimental samples and starts one timer when the last sample is treated. | ||
| − | + | [[Image:Fa22 M1D4 intro image.png|thumb|right|500px| Representative sampling is important for accurate data analysis.]]While some forms of bias, like the examples listed above, are easily mitigated (or might not have a significant effect at all), subconscious forms of bias are often the most sinister – and the most dangerous for the integrity of your experiment. This is especially relevant for image analysis. | |
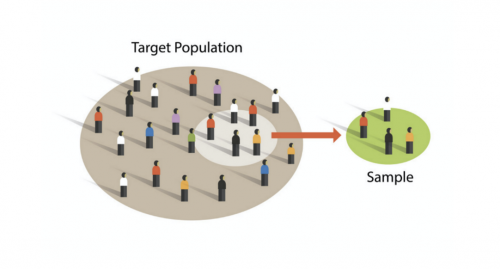
| − | + | When imaging, it is important to avoid selection bias to ensure that the sample appropriately represents the target population (see image to the right). Selection bias prevents a sample from being representative of the overall population. Say you have a condition that you believe should make cells more fluorescent in response to a reporter. How will you prevent yourself from leaning towards picking fields of view that are more fluorescent in your treatment group over the control group? When the instructors acquired images of your coverslips, the following criteria were used to mitigate selection bias. | |
| − | + | #The instructors used the DAPI channel to look for fields of view, NOT the γH2AX channel. | |
| − | + | #The first image was acquired by looking around the center of the coverslip for the first field of view that contained at least 5 cells | |
| − | + | #The second image was acquired by moving the field of view left until none of the cells in the first field of view were visible and until the field of view had at least 15 cells (when possible) | |
| − | + | #The third image was acquired in the same way as the second. | |
| − | # | + | |
| − | + | ||
| − | # | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | # | + | |
| − | + | ||
| − | + | ||
| − | # | + | |
| − | + | ||
| − | + | ==Protocols== | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ===Part 1: Analyze γH2AX images by counting foci=== | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | To analyze your data, you will use ImageJ and a plugin written by Joshua Corrigan to enumerate the γH2AX foci present in the nuclei of the treated cells. If you would like to review concepts used in this code, please review a protocol written by researchers at Duke University outlined [https://microscopy.duke.edu/guides/count-nuclear-foci-ImageJ here]. | |
| − | + | In this analysis, you will calculate the average foci per nuclei, indicating DNA strand breaks (γH2AX staining). Please note that this process is not automated, so each image will have to be analyzed individually. | |
| − | + | #To begin, ensure that all of your H2AX images are in an easily accessible location. The raw data can be found [https://www.dropbox.com/sh/c6wtogv13iioo9b/AAD-_4X_Z6n9MOQEjPe7NVfua?dl=0 here] and on the class data page. | |
| − | In this analysis, you will calculate the average foci per nuclei, indicating | + | #Download the quantification plugin [[Media:H2AX Quantification for MEFs.ijm|here]]. |
| − | #To begin, ensure that all of your H2AX images are in | + | #Open ImageJ and go to Analyze -> Set Measurements |
| − | #Download | + | #*In the Set Measurements window, make sure the following boxes are checked: Area, Mean gray value, Min & max gray value, Shape descriptors, Integrated density, Display label |
| − | # | + | #Go to File --> Open to open the macro window or drag the downloaded plugin into ImageJ. |
| − | # | + | #Open an image in ImageJ using the same approach as above. |
| − | # | + | #Click on the "Run" button in the macro window to begin analysis. |
| − | #* | + | #*This will separate the stacks of the multichannel image and allow you to begin the thresholding process. |
| − | # | + | #Manually set the threshold so that the background is black and the nuclei are solid red shapes reflecting the DAPI signal parameters in the image. |
| − | # | + | #*Make sure the cell nuclei are highlighted in red. |
| − | # | + | #Adjust the threshold values to properly identify the majority of the cells' nuclei. |
| − | # | + | [[Image:Raw maxima.png|thumb|right|250px|'''Match foci counts to image''']] |
| − | # | + | #Click "ok" to begin the process of watershedding the nuclei (defining the boundaries and separating nuclei in close proximity). The macro will also use the presence of the DAPI signal to create a "mask" for use in the FITC channel. |
| − | # | + | #*The program will switch to the image of the FITC channel to examine γH2AX sites. It will also turn up the brightness in this channel to allow foci to be more easily identified. |
| − | # | + | #You will now use the FITC channel to set a threshold level to identify foci. |
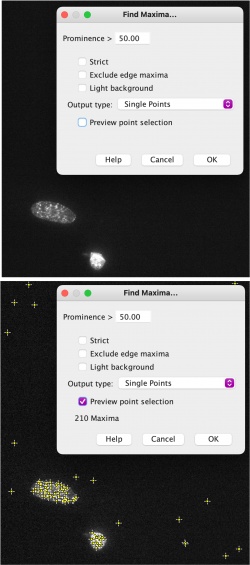
| − | # | + | #In the new window you will have the opportunity to set a prominence threshold. |
| − | #* | + | #Under the "output type" dropdown menu, select the "single points" option. |
| − | #* | + | #Check the box to preview point selection. |
| − | #* | + | #You will set the prominence so that the enhanced foci are counted, but the background signal is minimized. |
| − | # | + | #*Begin by setting the prominence at 500. Then check and uncheck the "preview points" box so that you are able to toggle back and forth between the raw image and processed one. |
| − | #* | + | #*Your goal is to see all foci in the image tagged with a plus sign to represent that the foci will be counted. |
| − | # | + | #**Record the prominence number you have chosen and use it for each subsequent image. |
| + | #When you have finished setting the parameters click OK to pull up the results sheet. | ||
| + | #Copy the results from the ImageJ results sheet into Excel. | ||
| + | #*DO NOT press 'ok' in ImageJ until the results have been copied as this will close your images. | ||
| + | #You will use the Excel files to assess how foci are identified in each nuclei. To do this, divide the Raw Integrated Density (RawIntDen) for each nuclei by 255 (each maxima representing a foci has a value of 255) to identify the number of foci in each nucleus. You can now average the number for each image in the condition. | ||
#*Repeat these steps for the additional conditions to quantify your results. | #*Repeat these steps for the additional conditions to quantify your results. | ||
| − | |||
<font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
| − | * | + | *Why did the instructors choose to select fields of view based on the DAPI channel rather than the yH2AX channel? |
| − | * | + | *Another potential way to analyze this data is to assess the global 488nm signal intensity in each nucleus using an automated macro and the same threshold applied to all images. Are there any advantages or disadvantages when comparing this method to the one you have used? |
| + | *What are two limitations to your current method? How would you address these limitations in future experiments? | ||
| − | ===Part | + | ===Part 2: Participate in group paper discussion=== |
To further help you in preparing your Data summary, we will discuss how similar data are presented in a publication from the Engelward laboratory. | To further help you in preparing your Data summary, we will discuss how similar data are presented in a publication from the Engelward laboratory. | ||
| Line 146: | Line 91: | ||
<font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
*Based on your reading and the group discussion of the article, answer the questions above. | *Based on your reading and the group discussion of the article, answer the questions above. | ||
| + | |||
| + | ===Part 3: Learn about the CometChip=== | ||
| + | |||
| + | As discussed in the prelab lecture, you will use two methods to assess DNA damage: the γH2AX assay and the CometChip. Before we review the CometChip experimental details, it is best to familiarize yourself with the procedure and assay. | ||
| + | |||
| + | Read the Abstract and Introduction in the following publication: | ||
| + | |||
| + | [[Media:CometChip Jove article.pdf| CometChip: A high-throughput 96-well platform for measuring DNA damage in microarrayed human cells.]] ''Journal of Visualized Experiments.'' (2014) 92: 1-11. | ||
| + | |||
| + | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> answer the following questions: | ||
| + | *Why is it important to study DNA damage? | ||
| + | **You can consider the information provided in lecture / prelab to answer this question. | ||
| + | *How does the CometChip estimate the level of DNA damage within cells? | ||
| + | *List two issues / problems with the comet assay? | ||
| + | *List two improvements provided by the CometChip assay? | ||
| + | |||
| + | ===Part 4: Prepare CometChip=== | ||
| + | |||
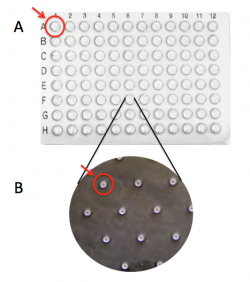
| + | [[Image:Fa18 M1D2 macrowell vs microwell.png|thumb|250px|right|'''Image of CometChip''' showing distinction between (A) 'macrowell' and (B) 'microwell'.]]The CometChip is simply a thin layer of agarose with microwells. It is important to differentiate between the terms 'macrowell' and 'microwell' for your experiments. A bottomless 96-well plate is placed on top of the agarose CometChip to create the macrowells for the CometChip assay (panel A). This enables researchers to control which cells are exposed to which treatment. The microwells were stamped into the agarose when you made your CometChip (panel B). Within each well are ~ 300 microwells, which are ~40 μm in diameter and 40 μm in depth. | ||
| + | |||
| + | <font color = #0d368e>'''To ensure the steps required for preparing a CometChip are clear, the Instructor will provide a live demonstration of this process. </font color> | ||
| + | |||
| + | #Obtain a sheet of gelbond film from the laboratory bench at the front of the room. The paper is protecting the hydrophilic side of the gelbond film. | ||
| + | #*Be sure to keep the paper associated with the gelbond film so you know which side is which. | ||
| + | #Prepare 20 mL of 1% normal melting point (NMP) agarose. '''Be careful as the agarose solution will be very hot!''' | ||
| + | #*Calculate the amount of NMP agarose powder needed for a 1% w/v solution. Check your math with the teaching faculty before you continue. | ||
| + | #*Obtain a small milk bottle from the front bench. | ||
| + | #*Weigh out the appropriate amount of NMP agarose and add it to the milk bottle. | ||
| + | #*Use a cylinder to measure 20 mL of 1x PBS and add it to the milk bottle with the NMP agarose powder. | ||
| + | #*Swirl to mix. | ||
| + | #*To melt the NMP agarose, microwave the solution for 20 seconds, swirl, then microwave for 3-second intervals until all crystals are in solutions. After each interval, remove the milk bottle and gently swirl while checking for unmelted agarose crystals. '''It is important that the solution does NOT boil as you will lose water to evaporation and the density of the agarose will be altered. If your solution starts to boil, immediately remove it from the microwave and gently swirl.''' | ||
| + | #*When no more crystals are visible in the solution take the milk bottle to your bench. | ||
| + | #Obtain a small rectangle dish (labeled "scraped lid") and the CometChip 'stamp' from the front bench. | ||
| + | #Add 2.5 mL of the agarose solution to the small dish, then ''quickly'' place the gelbond film in the dish with the marked hydrophobic side down. Remove the paper from the film. | ||
| + | #Add 13 mL of the agarose solution on top of the gelbond film. | ||
| + | #Slowly place the CometChip stamp on top of the agarose. | ||
| + | #*Lower the bottom left of the stamp first, then slowly allow the stamp to 'roll' into the agarose. Be sure to leave the top right corner of the small dish accessible. | ||
| + | #*Be careful not to introduce bubbles into the agarose and work quickly as the agarose will solidify as it cools. | ||
| + | #Allow the agarose to solidify, undisturbed, on your bench for 30 min. | ||
| + | #Add ~5 mL of 1x PBS to the small dish that contains your agarose CometChip. | ||
| + | #*Pipet in the 1x PBS using the accessible corner. | ||
| + | #Slowly pull from one corner of the stamp to lift it away from your CometChip in the dish. | ||
| + | #*If the CometChip sticks to the stamp, carefully peel it off using tweezers. | ||
| + | #*Discard the PBS in the sink. | ||
| + | #Remove excess agarose from the perimeter of your CometChip using a razor blade (obtain and return razor blade to front bench). | ||
| + | #Clean the agarose from the bottom of your CometChip (gelbond side) using a Kimwipe. | ||
| + | #Place your CometChip in the small dish containing 1x PBS for storage at 4 °C until next time. | ||
| + | #*Be sure the chip is completely submerged. | ||
| + | #Please return the stamp to the front bench. Never wipe the stamp as that will ruin the microposts! | ||
| + | |||
| + | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
| + | *Onto which side of the GelBond is the agarose poured to make a CometChip? | ||
| + | *How are the microwells generated in the agarose of the CometChip? | ||
==Reagents list== | ==Reagents list== | ||
| + | *agar, normal melting point (from Invitrogen) | ||
| + | *GelBond film (from Lonza) | ||
==Navigation links== | ==Navigation links== | ||
Next day: [[20.109(F22):M1D5 | Treat cells for CometChip assay]]<br> | Next day: [[20.109(F22):M1D5 | Treat cells for CometChip assay]]<br> | ||
Previous day: [[20.109(F22):M1D3 | Use immunoflourescence staining to assess γH2AX experiment]]<br> | Previous day: [[20.109(F22):M1D3 | Use immunoflourescence staining to assess γH2AX experiment]]<br> | ||
Latest revision as of 17:31, 27 September 2022
Contents
Introduction
Today you will analyze data from your first experiment in 20.109! When collecting data from an experiment, it is important to be a neutral observer. This will prevent you from introducing bias into the results. Bias refers to any treatment that differs between experimental groups that does NOT relate to the experimental question. To maximize reliability of data, scientists are trained to limit experimental bias as much as possible. To give you an idea of some of the many ways bias can be introduced into an experiment, consider the following scenarios:
- An experimenter uses the last remaining volume of buffer on their control samples. They make a new, fresh batch of buffer for their experimental samples.
- An experimenter always loads in their control samples and experimental samples in the same order into a 96 well plate when using an automated sampling machine.
- An experimenter treats all of their control samples, then all of their experimental samples and starts one timer when the last sample is treated.
When imaging, it is important to avoid selection bias to ensure that the sample appropriately represents the target population (see image to the right). Selection bias prevents a sample from being representative of the overall population. Say you have a condition that you believe should make cells more fluorescent in response to a reporter. How will you prevent yourself from leaning towards picking fields of view that are more fluorescent in your treatment group over the control group? When the instructors acquired images of your coverslips, the following criteria were used to mitigate selection bias.
- The instructors used the DAPI channel to look for fields of view, NOT the γH2AX channel.
- The first image was acquired by looking around the center of the coverslip for the first field of view that contained at least 5 cells
- The second image was acquired by moving the field of view left until none of the cells in the first field of view were visible and until the field of view had at least 15 cells (when possible)
- The third image was acquired in the same way as the second.
Protocols
Part 1: Analyze γH2AX images by counting foci
To analyze your data, you will use ImageJ and a plugin written by Joshua Corrigan to enumerate the γH2AX foci present in the nuclei of the treated cells. If you would like to review concepts used in this code, please review a protocol written by researchers at Duke University outlined here.
In this analysis, you will calculate the average foci per nuclei, indicating DNA strand breaks (γH2AX staining). Please note that this process is not automated, so each image will have to be analyzed individually.
- To begin, ensure that all of your H2AX images are in an easily accessible location. The raw data can be found here and on the class data page.
- Download the quantification plugin here.
- Open ImageJ and go to Analyze -> Set Measurements
- In the Set Measurements window, make sure the following boxes are checked: Area, Mean gray value, Min & max gray value, Shape descriptors, Integrated density, Display label
- Go to File --> Open to open the macro window or drag the downloaded plugin into ImageJ.
- Open an image in ImageJ using the same approach as above.
- Click on the "Run" button in the macro window to begin analysis.
- This will separate the stacks of the multichannel image and allow you to begin the thresholding process.
- Manually set the threshold so that the background is black and the nuclei are solid red shapes reflecting the DAPI signal parameters in the image.
- Make sure the cell nuclei are highlighted in red.
- Adjust the threshold values to properly identify the majority of the cells' nuclei.
- Click "ok" to begin the process of watershedding the nuclei (defining the boundaries and separating nuclei in close proximity). The macro will also use the presence of the DAPI signal to create a "mask" for use in the FITC channel.
- The program will switch to the image of the FITC channel to examine γH2AX sites. It will also turn up the brightness in this channel to allow foci to be more easily identified.
- You will now use the FITC channel to set a threshold level to identify foci.
- In the new window you will have the opportunity to set a prominence threshold.
- Under the "output type" dropdown menu, select the "single points" option.
- Check the box to preview point selection.
- You will set the prominence so that the enhanced foci are counted, but the background signal is minimized.
- Begin by setting the prominence at 500. Then check and uncheck the "preview points" box so that you are able to toggle back and forth between the raw image and processed one.
- Your goal is to see all foci in the image tagged with a plus sign to represent that the foci will be counted.
- Record the prominence number you have chosen and use it for each subsequent image.
- When you have finished setting the parameters click OK to pull up the results sheet.
- Copy the results from the ImageJ results sheet into Excel.
- DO NOT press 'ok' in ImageJ until the results have been copied as this will close your images.
- You will use the Excel files to assess how foci are identified in each nuclei. To do this, divide the Raw Integrated Density (RawIntDen) for each nuclei by 255 (each maxima representing a foci has a value of 255) to identify the number of foci in each nucleus. You can now average the number for each image in the condition.
- Repeat these steps for the additional conditions to quantify your results.
In your laboratory notebook, complete the following:
- Why did the instructors choose to select fields of view based on the DAPI channel rather than the yH2AX channel?
- Another potential way to analyze this data is to assess the global 488nm signal intensity in each nucleus using an automated macro and the same threshold applied to all images. Are there any advantages or disadvantages when comparing this method to the one you have used?
- What are two limitations to your current method? How would you address these limitations in future experiments?
Part 2: Participate in group paper discussion
To further help you in preparing your Data summary, we will discuss how similar data are presented in a publication from the Engelward laboratory.
Weingeist, D. M., et al. "Single-cell microarray enables high-throughput evaulation of DNA double-strand breaks and DNA repair inhibitors." Cell Cycle. (2013) 126:907-915.
From the Introduction
Consider the key components of an introduction:
- What is the big picture?
- Is the importance of this research clear?
- Are you provided with the information you need to understand the research?
- Do the authors include a preview of the key results?
From the Results
Carefully examine the figures. First, read the captions and use the information to 'interpret' the data presented within the image. Second, read the text within the results section that describes the figure.
- Do you agree with the conclusion(s) reached by the authors?
- What controls were included and are they appropriate for the experiment performed?
- Are you convinced that the data are accurate and/or representative?
From the Discussion
Consider the following components of a discussion:
- Are the results summarized?
- Did the authors 'tie' the data together into a cohesive and well-interpreted story?
- Do the authors overreach when interpreting the data?
- Are the data linked back to the big picture from the introduction?
In your laboratory notebook, complete the following:
- Based on your reading and the group discussion of the article, answer the questions above.
Part 3: Learn about the CometChip
As discussed in the prelab lecture, you will use two methods to assess DNA damage: the γH2AX assay and the CometChip. Before we review the CometChip experimental details, it is best to familiarize yourself with the procedure and assay.
Read the Abstract and Introduction in the following publication:
CometChip: A high-throughput 96-well platform for measuring DNA damage in microarrayed human cells. Journal of Visualized Experiments. (2014) 92: 1-11.
In your laboratory notebook, answer the following questions:
- Why is it important to study DNA damage?
- You can consider the information provided in lecture / prelab to answer this question.
- How does the CometChip estimate the level of DNA damage within cells?
- List two issues / problems with the comet assay?
- List two improvements provided by the CometChip assay?
Part 4: Prepare CometChip
The CometChip is simply a thin layer of agarose with microwells. It is important to differentiate between the terms 'macrowell' and 'microwell' for your experiments. A bottomless 96-well plate is placed on top of the agarose CometChip to create the macrowells for the CometChip assay (panel A). This enables researchers to control which cells are exposed to which treatment. The microwells were stamped into the agarose when you made your CometChip (panel B). Within each well are ~ 300 microwells, which are ~40 μm in diameter and 40 μm in depth.To ensure the steps required for preparing a CometChip are clear, the Instructor will provide a live demonstration of this process.
- Obtain a sheet of gelbond film from the laboratory bench at the front of the room. The paper is protecting the hydrophilic side of the gelbond film.
- Be sure to keep the paper associated with the gelbond film so you know which side is which.
- Prepare 20 mL of 1% normal melting point (NMP) agarose. Be careful as the agarose solution will be very hot!
- Calculate the amount of NMP agarose powder needed for a 1% w/v solution. Check your math with the teaching faculty before you continue.
- Obtain a small milk bottle from the front bench.
- Weigh out the appropriate amount of NMP agarose and add it to the milk bottle.
- Use a cylinder to measure 20 mL of 1x PBS and add it to the milk bottle with the NMP agarose powder.
- Swirl to mix.
- To melt the NMP agarose, microwave the solution for 20 seconds, swirl, then microwave for 3-second intervals until all crystals are in solutions. After each interval, remove the milk bottle and gently swirl while checking for unmelted agarose crystals. It is important that the solution does NOT boil as you will lose water to evaporation and the density of the agarose will be altered. If your solution starts to boil, immediately remove it from the microwave and gently swirl.
- When no more crystals are visible in the solution take the milk bottle to your bench.
- Obtain a small rectangle dish (labeled "scraped lid") and the CometChip 'stamp' from the front bench.
- Add 2.5 mL of the agarose solution to the small dish, then quickly place the gelbond film in the dish with the marked hydrophobic side down. Remove the paper from the film.
- Add 13 mL of the agarose solution on top of the gelbond film.
- Slowly place the CometChip stamp on top of the agarose.
- Lower the bottom left of the stamp first, then slowly allow the stamp to 'roll' into the agarose. Be sure to leave the top right corner of the small dish accessible.
- Be careful not to introduce bubbles into the agarose and work quickly as the agarose will solidify as it cools.
- Allow the agarose to solidify, undisturbed, on your bench for 30 min.
- Add ~5 mL of 1x PBS to the small dish that contains your agarose CometChip.
- Pipet in the 1x PBS using the accessible corner.
- Slowly pull from one corner of the stamp to lift it away from your CometChip in the dish.
- If the CometChip sticks to the stamp, carefully peel it off using tweezers.
- Discard the PBS in the sink.
- Remove excess agarose from the perimeter of your CometChip using a razor blade (obtain and return razor blade to front bench).
- Clean the agarose from the bottom of your CometChip (gelbond side) using a Kimwipe.
- Place your CometChip in the small dish containing 1x PBS for storage at 4 °C until next time.
- Be sure the chip is completely submerged.
- Please return the stamp to the front bench. Never wipe the stamp as that will ruin the microposts!
In your laboratory notebook, complete the following:
- Onto which side of the GelBond is the agarose poured to make a CometChip?
- How are the microwells generated in the agarose of the CometChip?
Reagents list
- agar, normal melting point (from Invitrogen)
- GelBond film (from Lonza)
Next day: Treat cells for CometChip assay