Difference between revisions of "20.109(F21):M2D1"
Noreen Lyell (Talk | contribs) |
Noreen Lyell (Talk | contribs) (→Navigation links) |
||
| Line 18: | Line 18: | ||
==Navigation links== | ==Navigation links== | ||
Next day: [[20.109(F21):M2D2 |Complete in silico cloning of protein expression plasmid]] <br> | Next day: [[20.109(F21):M2D2 |Complete in silico cloning of protein expression plasmid]] <br> | ||
| − | Previous day | + | Previous day: [[20.109(F21):M1D7 | Analyze data using statistical methods]]<br> |
Revision as of 17:20, 2 September 2021
Introduction
A small molecule microarray (SMM) requires the covalent attachment of a library of small molecules to a glass slide. Our library is meant to broadly sample the drug-like chemical space (i.e. all possible chemical structures that have drug-like physical properties) and contains about 50,000 small molecules. Some libraries are much smaller, while many pharmaceutical companies possess high-throughput screening (HTS) collections of millions of compounds. Because this chemical space is very large, it’s difficult to generalize any single chemical reaction for this attachment that can be applied to all small molecules. We take a “one-size-fits-most” approach, where the glass slide is functionalized with a broadly reactive electrophile capable of reacting with nucleophiles present in most drug-like small molecules, such as alcohols or amines. Many small molecules contain multiple nucleophiles suitable for attachment. In this case, our manufacturing will result in a mixture of attachment sites. It’s important to remember that attachment to the glass slide constrains the possible orientation of the protein-small molecule interaction; some orientations are not possible because the glass slide and linker are in the way.
We start with a glass slide with exposed amines across the surface and attach a short PEG (polyethylene glycol) linker. To the end of this PEG linker, an isocyanate group is attached. Isocyanate, or R-N=C=O, is a resonant structure, and a partial positive charge is stabilized on the carbon atom. This carbon atom is electrophilic, and small molecules with nucleophiles will react here. We estimate that about 70% of drug-like small molecules are amenable to this reaction, and our library is filtered to contain only these molecules.
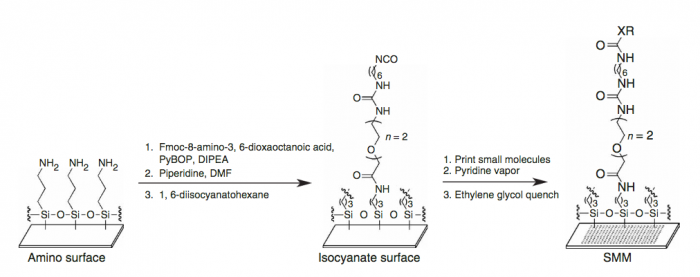
Our compound library is dissolved in DMSO and stored in 384-well plates. To dispense the compounds onto our functionalized glass slide, a robotic arm with a set of 48 metal pins is used to transfer the compounds to the glass slide. Each metal pin has a small slit in the end, and capillary action is used to precisely withdraw and dispense consistent volumes. When the pins touch the glass slide, the compound in DMSO is dispensed into a small circle of approximately 160 micron in diameter. Each pin prints one compound in two different locations on each slide, and then the pins are washed in water and DMSO. This process is repeated for each compound, resulting in our final microarray. The microarray is divided into 48 subarrays, and each subarray corresponds to one pin and contains 256 discrete spots. Within each subarray, we print a set of fluorescent compounds in the shape of an ‘X’ so that we can precisely determine where each spot is printed. After the compounds react, we quench the surface so that no electrophiles remain. This results in our final microarray; a collection of approximately 12,000 discrete spots displaying one compound each.
(Written by Rob Wilson. For more information read Bradner, J. E., McPherson, O. M., and Koehler, A. N. (2006) A method for the covalent capture and screening of diverse small molecules in a microarray format. Nature Prot. 1:2344-2352. PMID: 17406478.)
Protocols
Next day: Complete in silico cloning of protein expression plasmid