Difference between revisions of "20.109(F20):M3D3"
Noreen Lyell (Talk | contribs) (→Part 2: Analyze ethanol yield data) |
Noreen Lyell (Talk | contribs) (→Reagents list) |
||
| Line 126: | Line 126: | ||
==Reagents list== | ==Reagents list== | ||
| + | |||
| + | *Luria-Bertani (LB) broth: 1% tryptone, 0.5% yeast extract, and 1% NaCl | ||
| + | *chloramphenicol antibiotic stock (25 mg/mL) | ||
| + | *ampicillin antibiotic stock (100 mg/mL) | ||
| + | *anhydrotetracycline (aTc) stock (1 mM) (from Sigma Aldrich) | ||
| + | *Ethanol Assay Kit (from Sigma-Adrich) | ||
| + | **ethanol assay buffer | ||
| + | **ethanol probe | ||
| + | **ethanol enzyme | ||
==Navigation links== | ==Navigation links== | ||
Next day: [[20.109(F20):M3D4 | Examine genomic features in gRNA-targeted sequences]]<br> | Next day: [[20.109(F20):M3D4 | Examine genomic features in gRNA-targeted sequences]]<br> | ||
Previous day: [[20.109(F20):M3D2 | Align gRNA sequences with genomic targets]] | Previous day: [[20.109(F20):M3D2 | Align gRNA sequences with genomic targets]] | ||
Revision as of 01:49, 20 July 2020
Contents
Introduction
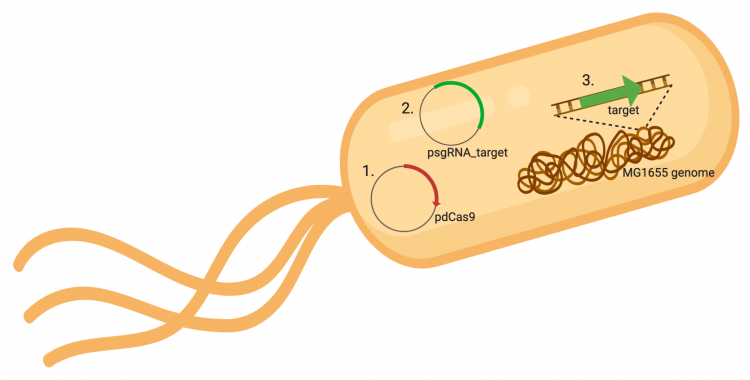
The CRISPRi system involves three genetic elements: the target gene within the host genome, the pdCas9 plasmid, and the pgRNA_target plasmid. Though the target genome is native to the host cell, the plasmids must be transformed into the cell and maintained using antibiotic selection. Throughout this module, we have learned about and worked with the CRISPRi plasmids as individual units, but now we will consider the system as a whole in the context of gene regulation.
In the previous laboratory session, you co-transformed pdCas9 and your pgRNA_target plasmids into E. coli MG1655. The colonies present on your LB plates containing chloramphenicol and ampicillin should carry both plasmids in that they were able to survive selection by both antibiotics. The promoter (pJ23119) driving expression of your gRNA sequence in the gRNA_target plasmid is constitutively active. This means that RNAP is not prohibiting from binding and transcription of the gRNA target sequence is therefore not inhibited. Therefore, your gRNA_target is always present in the MG1655 cells and binding to the target sequence within the genome.
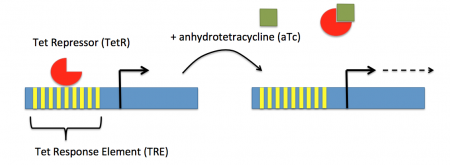
In contrast, expression of the gene encoding Cas9 within pdCas9 is regulated by an inducible promoter (pLtetO-1). An inducible promoter is 'off' unless the appropriate molecule is present to relieve repression. In the case of our system, expression of the gene encoding Cas9 is inhibited due to the use of a tet-based promoter construct. Tet is shorthand for tetracycline, which is an antibiotic that inhibits protein synthesis through preventing the association between charged aminoacyl-tRNA molecules and the A site of ribosomes. Bacterial cells that carry the tet resistance cassette are able to survive exposure to tetracycline by expressing genes that encode an efflux pump that 'flushes' the antibiotic from the bacterial cell. To conserve energy, the tet system is only expressed in the presence of tetracyline. In the absence of tetracycline, a transcription repressor protein (TetR) is bound to the promoter upstream of the tet resistance cassette genes. When tetracycline is present, the molecule binds to TetR causing a confirmational resulting in TetR 'falling off' of the promoter. In the CRISPRi system, the tet-based promoter construct upstream of the gene that encodes Cas9 is 'off' unless anhydrotetracyline (aTc), an analog of tetracyline, is added to the culture media.Taken together, the gRNA-target molecule is constitutively transcribed and, as stated above, always present / binding to the target. The dCas9 protein is only present when aTc is added. Thus, gene expression is only altered when aTc is present. As represented by the schematic below, the gRNA_target 'seeks out' the target within the host genome and recruits dCas9 to the site. When associated with the target / gRNA_target complex, dCas9 binds to the site and acts as a 'roadblock' by prohibiting RNAP access to the sequence. Because the gene of interest is not able to be transcribed, the protein encoded by that gene is not synthesized. In our experiments, we hypothesize that the absence of specific proteins, or enzymes, in the fermentation pathway will increase the yield of either ethanol or acetate.
Protocols
Part 1: Review experimental setup
Prepare media for mixed-acid fermentation inoculations
To test the effect of your gRNA on altering the production of either ethanol or lactate, you will incubate the co-transformed E. coli cells both with and without oxygen. Remember from lecture that cells grown anaerobically will produce more fermentation products; however, our goal is to further enhance the production of these products by manipulating the fermentation pathway.
- Acquire 4 glass culture tubes and 4 15 mL conical tubes from the front laboratory bench and label as shown below:
- MG1655 +O2 -aTc (glass tube)
- MG1655 -O2 -aTc (15 mL conical tube)
- MG1655 +O2 +aTc (glass tube)
- MG1655 -O2 +aTc (15 mL conical tube)
- MG1655 +CRISPRi +O2 -aTc (glass tube)
- MG1655 +CRISPRi -O2 -aTc (15 mL conical tube)
- MG1655 +CRISPRi +O2 +aTc (glass tube)
- MG1655 +CRISPRi -O2 +aTc (15 mL conical tube)
- Be sure to include your team information on each tube!
- Obtain a bottle of LB broth from the front laboratory bench.
- Transfer 5 mL of the media into each tube labeled for inoculation with MG1655 (alone, not co-transformed).
- Calculate the volume of the chloramphenicol and ampicillin antibiotic stocks that are required to maintain the pdCas9 and pgRNA plasmids, respectively, in the remaining volume of the LB media using the information below:
- For chloramphenicol: the stock concentration is 25 mg/mL and the final or 'working' concentration should be 25 μg/mL.
- For ampicillin: the stock concentration is 100 mg/mL and the final or 'working' concentration should be 100 μg/mL.
- Add the appropriate volume of each antibiotic stock to the LB aliquot and mix thoroughly.
- Transfer 5 mL of the media you prepared in Step #5 into each tube labeled for inoculation with MG1655 +CRISPRi.
On either Monday or Tuesday afternoon, depending on your laboratory section, the teaching faculty will inoculate MG1655 or a co-transformant into your culture tubes and add 2 μM aTc (final concentration) to the appropriate tubes. All tubes will then be incubated at 37 °C until you return for the next laboratory session.
Measure cell density of cultures
Before you prepare your samples to quantify the fermentation product yield, it is important that you measure the optical density (OD) of your cultures. This number will be used to normalize your data, which allows you to more accurately compare your samples. As a reminder, the OD is a measure of culture density and is based on the deflection of light at 600 nm (OD600). In this method of analysis, the higher the OD is the more dense the culture.
- Obtain an aliquot of fresh LB media and 9 plastic cuvettes from the front laboratory bench.
- Pipet 1000 μL of LB into 1 cuvette. This will serve as the blank needed to calibrate the spectrophotometer.
- Pipet 900 μL of LB into each of the 8 remaining cuvettes.
- It may be helpful to label your cuvettes with a simple designation that corresponds to your samples. Be sure that your labels are not in the light path as this will obscure your OD600 readings.
- Transfer 100 μL of each culture into the appropriate plastic cuvette.
- Note: this is a 1:10 dilution of your sample that should be accounted for in your calculations for product yield.
- Use the spectrophotometer to measure the OD600 values according to the instructions from the Orientation exercise.
- This step will be a bottleneck. Please be courteous to your laboratory mates and do not go to the spectrophotometer until your samples are fully prepared and ready to be measured.
Quantify fermentation product yield
Please be sure to follow the appropriate protocol below.
Measuring ethanol yield
Prepare supernatants from overnight cultures
- Transfer the cultures from your aerobic tubes to fresh 15 mL conical tubes.
- Be sure to label your tubes!
- Pellet the bacterial cells in your cultures by centrifugation using the large centrifuge on the teaching faculty laboratory bench.
- Alert the teaching faculty when you are ready for centrifugation.
- Centrifuge your samples at 3000 rpm for 10 min.
- If the media is still ‘cloudy’ repeat the centrifugation step.
- Pipet 1000 μL of the supernatant from each 15 mL conical tube into a fresh 1.5 mL eppendorf tube.
- Be sure to label your tubes!
- Label 8 additional eppendorf tubes and generate 1:15 dilutions of each of your samples in a final volume of 300 μL.
- Use fresh LB as the diluent.
Prepare samples for standard curve
- Prepare a 1 nmole/μL stock ethanol solution:
- In an eppendorf tube labeled A, combine 808.7 μL of the Ethanol Assay Buffer with 50 μL of the 17.15 N Ethanol Standard to generate a 1 μmole/μL solution.
- In an eppendorf tube labeled B, combine 990 μL of Ethanol Assay Buffer with 10 μL of solution A (from tube A) to generate a 10 nmole/μL solution.
- In an eppendorf tube labeled C, combine 900 μL of Ethanol Assay Buffer with 100 μL of solution B (from tube B) to generate a 1 nmole/μL stock ethanol solution.
- Label 5 eppendorf tubes as follows: 2 nmole, 4 nmole, 6 nmole, 8 nmole, and 10 nmole. (These will be the amounts in 50 μL samples, as will make sense in a few minutes.) Use the following steps to prepare samples for your standard curve that contain the specified amounts of ethanol.
- In the 2 nmole tube, combine 144 μL of the Ethanol Assay Buffer with 6 μL of the 1 nmole/μL stock ethanol solution you generated in Step #1.
- In the 4 nmole tube, combine 138 μL of the Ethanol Assay Buffer with 12 μL of the 1 nmole/μL stock ethanol solution you generated in Step #1.
- Prepare the 6 nmole, 8 nmole, and 10 nmole tubes.
- Note: the amount of ethanol in each tube is three times as much as specified by the label because you are preparing enough for duplicate samples (ie the contents of each tube will be divided between two wells in your assay resulting in the appropriate amount of ethanol in each well).
Complete ethanol assay
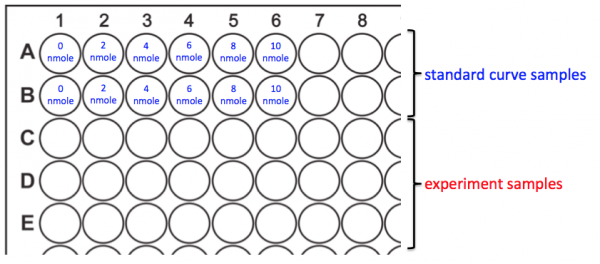
- You will prepare your assay plate according to the map above.
- Add 50 μL of the Ethanol Assay Buffer to wells A1 and B1. This is the 0 nmole/well sample for your standard curve.
- Add 50 μL of each of the remaining standard curve samples to the appropriate wells.
- Add 50 μL of your experimental samples according to the following order:
- To C1, D1 and E1 add MG1655 +O2 -aTc
- To C2, D2 and E2 add MG1655 -O2 -aTc
- To C3, D3 and E3 add MG1655 +O2 +aTc
- To C4, D4 and E4 add MG1655 -O2 +aTc
- To C5, D5 and E5 add MG1655 +CRISPRi +O2 -aTc
- To C6, D6 and E6 add MG1655 +CRISPRi -O2 -aTc
- To C7, D7 and E7 add MG1655 +CRISPRi +O2 +aTc
- To C8, D8 and E8 add MG1655 +CRISPRi -O2 +aTc
- Calculate the amount of Reaction Mix you will need for your assay using the following information:
- To account for pipetting error, you will add an additional 20% of each component to the mixture.
- You will add 50 μL of Reaction Mix to every well (supernatants and standard curve samples).
- The Reaction Mix consists of 46 μL of Ethanol Assay Buffer, 2 μL of Ethanol Probe, and 2 μL of Ethanol Enzyme Mix per 50 μL.
- Confirm your calculations with the teaching faculty before you proceed.
- Combine the appropriate amount of Ethanol Assay Buffer, Ethanol Probe, and Ethanol Enzyme Mix in a fresh, labeled conical tube.
- Add 50 μL of your Reaction Mix to each well in your 96-well plate that contains either standard or experimental samples.
- After each addition, use the pipet to mix the contents of the well.
- Be sure to change pipet tips between additions.
- Cover your 96-well plate tightly with aluminum foil and incubate at room temperature for 60 min.
- Alert the teaching faculty when your samples are ready for the spectrophotometer.
- Because the laboratory spectrophotometer is only able to measure a single sample in a cuvette, we will use a plate reader spectrophotometerto measure the A570 values for your assay.
Part 2: Analyze ethanol yield data
- Prepare a spreadsheet with the spectrophotometer values obtained for your assay.
- Average the technical replicates in your data set.
- Correct for the background ‘noise’ in your data by subtracting the averaged value of your 0 nmole/μL samples from the averaged values of all other samples.
- Plot the corrected values for your standard curve samples with the known concentration on the y-axis and the A570 (for ethanol) or A450 (for acetate) on the x-axis.
- Include the R-squared value and the equation of the best-fit line on your graph.
- Attach the graph to your Benchling laboratory notebook entry.
- Use the equation of the best-fit line to calculate the amount of fermentation produce in your experimental samples.
- For the ethanol and acetate assay: remember that you used a dilution of the supernatant in your assay!
- To calculate the concentration of product, use the following equation: Sa / Sv = C
- Sa = amount of fermentation product in unknown sample (nmole) from standard curve
- Sv = sample volume (μL) added to well
- C = concentration of fermentation product in sample
- The units of this calculation will be nmole/μL. For units of ng/μL use the molecular weight value of ethanol, which is 46.07 g/mol, or acetate, which is 60.05 g/mol.
- Lastly, normalize your data to the OD600 of your initial culture to account for difference in cell number.
- Divide the amount or concentration of fermentation produced by the OD value measured.
Reagents list
- Luria-Bertani (LB) broth: 1% tryptone, 0.5% yeast extract, and 1% NaCl
- chloramphenicol antibiotic stock (25 mg/mL)
- ampicillin antibiotic stock (100 mg/mL)
- anhydrotetracycline (aTc) stock (1 mM) (from Sigma Aldrich)
- Ethanol Assay Kit (from Sigma-Adrich)
- ethanol assay buffer
- ethanol probe
- ethanol enzyme
Next day: Examine genomic features in gRNA-targeted sequences