20.109(F20):M3D1
Contents
Introduction
In this module you build on the progress of former 109ers! In previous semesters, students engineered the the metabolism of E. coli in an attempt to increase the production of a commercially relevant product. Specifically, metabolic engineering was used to increase the ethanol yield via manipulating the fermentation pathway native to E. coli cells. Ethanol, or ethyl alcohol, is used as fuel – most commonly as a biofuel additive in gasoline. Current methods for producing ethanol involve the use of agricultural feedstocks and concern is mounting given the effect this may have on food prices and resources. Using E. coli as an ethanol-generating machine is promising; however, the ethanol output of a native cell is very low.
Metabolic engineering refers to the alteration of genetic and/or regulatory circuitry within organisms. The native circuitry involves a series of enzymes that perform biochemical reactions that function to convert raw substrates into products that are required for the organism’s survival. When this native circuitry is altered using metabolic engineering techniques, the goal is to use the host organisms as machines that produce valuable materials in large quantities and at low cost.
Common strategies employed in metabolic engineering are:- Increasing expression of a gene that encodes an enzyme responsible for a rate-limiting step.
- Inhibiting competing pathways that divert substrate away from the pathway of interest.
- Incorporating genes from other organisms into the host.
- Altering the structure of the enzyme such that yield is increased.
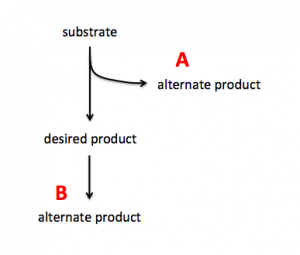
To increase production of a desired product, you can either target proteins that use the substrate to generate alternate products (see A in image on right) or you can target proteins that use the desired product to generate alternate products (see B in image on right). In A the substrate is siphoned away from the reaction that generates the desired product and in B the desired product is used as substrate in a subsequent reaction. By eliminating the proteins that catalyze the reactions that result in alternate products, you can potentially increase production of your desired product.
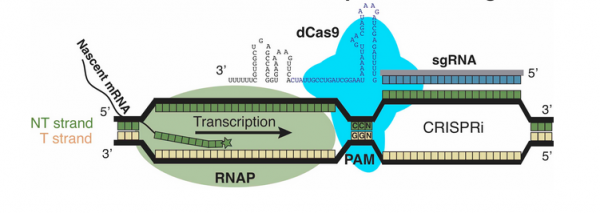
To manipulate the metabolism of E. coli, the CRISPRi system was used. Unlike more commonly used CRISPR-based technologies, CRISPRi is used to modulate expression from the genome rather than to modify the genome. This distinction is due to the use of an enzymatically-inactive dCas9 (or ‘dead’ Cas9) protein. Because dCas9 is enzymatically inactive, it is unable to cleave the DNA upon binding to a gRNA/DNA complex. The lack of DNA cleavage results in gene silencing through impeding RNA polymerase binding, transcription factor binding, and/or transcription elongation. The method of repression is largely due to the location of the genome that is targeted.
Protocols
The focus for today is to understand the key concepts and methods that were used to generate the data you will critically evaluate in this module. First, it is important to consider how targets were selected for the metabolic engineering approach. Second, you will review the CRISPRi technology used for the metabolic engineering approach.
Part 1: Review E. coli ethanol metabolism
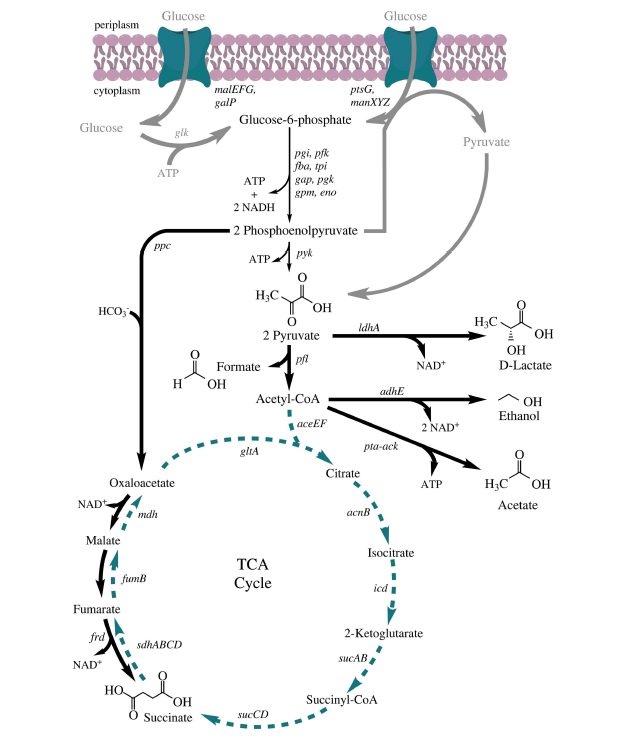
In E. coli ethanol is a product of the mixed-acid fermentation pathway. You will complete this task using the CRISPRi system, which targets genes such that transcription of the target gene is decreased, thus resulting in less of the protein encoded by the targeted gene.
With your laboratory partner, review Metabolic engineering of Escherichia coli for production of mixed-acid fermentation end products by Forster & Gescher. Using the information within this article and the genes that you identified above, select one gene that you will target in an attempt to increase the production of either ethanol or acetate. Be sure to include notes on your decision and thought process in your laboratory notebook.
Use the fermentation pathway from E. coli (included below) and the example above to answer the following questions with your partner.
- Which gene(s) might you target to increase the availability of substrate for ethanol production? For acetate production?
- Which gene(s) might you target to decrease the amount of substrate used to generate products in steps downstream of ethanol production? of acetate production?
===Part 2: Learn about CRISPRi system
Part 2: Practice gRNA design principles
WRITE EXERCISE USING REFERENCE PAPER AND JOSEPHINES PRACTICE SHEET
Part 3: Consider gRNA expression plasmid construction strategy
PCR amplification
The applications of PCR (polymerase chain reaction) are widespread, from forensics to molecular biology to evolution, but the goal of any PCR is the same: to generate many copies of DNA from a single or a few specific sequence(s) (called the “target” or “template”).
In addition to the target, PCR requires only three components: primers to bind sequence flanking the target, dNTPs to polymerize, and a heat-stable polymerase to carry out the synthesis reaction over and over and over. DNA polymerases require short initating pieces of DNA (or RNA) called primers in order to copy DNA. In PCR amplification, forward and reverse primers that target the non-coding and coding strands of DNA, respectively, are separated by a distance equal to the length of the DNA to be copied. Length is one important design feature. Primers that are too short may lack requisite specificity for the desired sequence, and thus amplify an unrelated sequence. The longer a primer is, the more favorable are its energetics for annealing to the template DNA, due to increased hydrogen bonding. On the other hand, longer primers are more likely to form secondary structures such as hairpins, leading to inefficient template priming. Two other important features are G/C content and placement. Having a G or C base at the end of each primer increases priming efficiency, due to the greater energy of a GC pair compared to an AT pair. The latter decrease the stability of the primer-template complex. Overall G/C content should ideally be 50 +/- 10%, because long stretches of G/C or A/T bases are both difficult to copy. The G/C content also affects the melting temperature. PCR is a three-step process (denature, anneal, extend) and these steps are repeated 20 or more times. After 30 cycles of PCR, there could be as many as a billion copies of the original target sequence.
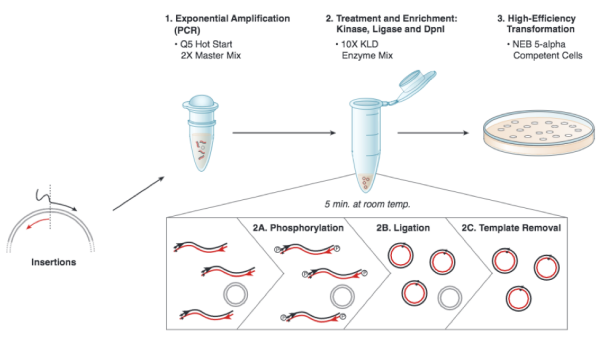
Based on the numerous applications of PCR, it may seem that the technique has been around forever. In fact it is just over 30 years old. In 1984, Kary Mullis described this technique for amplifying DNA of known or unknown sequence, realizing immediately the significance of his insight. We will be using the Q5 Site Directed Mutagenesis Kit from NEB to insert the gRNA sequence into an expression vector. For this procedure you will combine the gRNA primer you designed, a universal CRISPRi primer specific to pgRNA, and the pgRNA plasmid DNA encoding. DNA polymerase will copy the plasmid using the gRNA primer to insert the target sequence you selected. Following this reaction the 'mutated' product is a linear DNA fragment. To generate circular plasmids that carry the gRNA sequence, the DNA is phosphorylated then ligated. In addition, there is still parental -- that is, non-mutant -- DNA present in your reaction product. To ensure that only the gRNA-containing plasmid is used in the next steps, the parental DNA is selectively digested using the DpnI enzyme. The underlying selective property is that DpnI only digests methylated DNA. Therefore, the synthetically made (and thus non-methylated) mutant DNA is not digested, while the parental DNA is digested due to methylation by the host bacterial strain originally used to amplify it. The resulting small linear pieces of parental DNA are simply degraded by the bacteria upon transformation, whereas the intact (due to the phosphorylation and ligation reaction) circular mutant DNA is amplified by the bacteria.
Each group will set up one reaction, for your insertion. Meanwhile, the teaching faculty will set up a single positive control reaction, to ensure that all the reagents are working properly. You should work quickly but carefully, and keep your tube in a chilled container at all times. Please return shared reagents to the ice bucket(s) from which you took them as soon as you are done with each one.
- Get a PCR tube and label the top with your team color and lab section (write small!).
- Add 10.25 μL of nuclease-free water.
- Add 1.25 μL of your primer mix (each primer should be at a concentration of 10 μM).
- Add 1 μL of pgRNA plasmid DNA (concentration of 25 ng/μL).
- Lastly, use a filter tip to add 12.5 μL of Q5 Hot Start High-Fidelity 2X Master Mix - containing buffer, dNTPs, and polymerase - to your tube.
- Once all groups are ready, we will begin the thermocycler, under the following conditions:
| Segment | Cycles | Temperature | Time |
|---|---|---|---|
| Initial denaturation | 1 | 98 °C | 30 s |
| Amplification | 25 | 98 °C | 10 s |
| 55 °C | 30 s | ||
| 72 °C | 2 min | ||
| Final extension | 1 | 72 °C | 2 min |
| Hold | 1 | 4 °C | indefinite |
- After the cycling is completed, you will complete the KLD reaction (which stands for "kinase, ligase, DpnI") using
- 1 μL of your amplification product
- 5 μL 2X KLD Reaction Buffer
- 1 μL KLD Enzyme Mix, and
- 3 μL nuclease-free water.
- Incubate the reaction for 5 min at room temperature.
- Then, use 5 μL of the KLD reaction product to complete a transformation into an E. coli strain (NEB 5α cells of genotype fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17) that will amplify the plasmid such that you are able to confirm the appropriate insertion (or 'mutation') was incorporated. The transformation procedure will be as follows:
- Add 5 μL of KLD mix to 50 μL of chemically-competent NEB 5α.
- Incubate on ice for 30 min.
- Heat shock at 42 °C for 30 s.
- Incubate on ice for 5 min.
- Add 950 μL SOC and gently shake at 37 °C for 1 h.
- Spread 50 μL onto LB+Amp plate and incubate overnight at 37 °C.
Reagents list
- Q5 Site Directed Mutagenesis Kit (from NEB)
- Q5 Hot Start High-Fidelity 2X Master Mix: propriety mix of Q5 Hot Start High-Fidelity DNA Polymerase, buffer, dNTPs, and Mg2+
- 2X KLD Reaction Buffer
- 10X KLD Enzyme Mix: proprietary mix of kinase, ligase, and DpnI enzymes
- Universal CRIPSRi reverse primer 5' - ACT AGT ATT ATA CCT AGG ACT GAG CTA GC - 3'
- SOC medium: 2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose
- LB+Amp plates
- Luria-Bertani (LB) broth: 1% tryptone, 0.5% yeast extract, and 1% NaCl
- Plates prepared by adding 1.5% agar and 100 μg/mL ampicillin (Amp) to LB
Next day: Align gRNA sequences with genomic targets