Difference between revisions of "20.109(F20):M3D1"
Noreen Lyell (Talk | contribs) |
Noreen Lyell (Talk | contribs) (→Part 2: Research CRISPRi system) |
||
| (40 intermediate revisions by one user not shown) | |||
| Line 4: | Line 4: | ||
==Introduction== | ==Introduction== | ||
| − | In this module you build | + | In this module you will build upon the progress of former 109ers! In previous semesters, students engineered the the metabolism of ''E. coli'' in an attempt to increase the production of a commercially relevant product. Specifically, metabolic engineering was used to increase the ethanol yield via manipulating the fermentation pathway native to ''E. coli'' cells. Ethanol, or ethyl alcohol, is used as fuel – most commonly as a biofuel additive in gasoline. Current methods for producing ethanol involve the use of agricultural feedstocks and concern is mounting given the effect this may have on food prices and resources. Using ''E. coli'' as an ethanol-generating machine is promising; however, the ethanol output of a native cell is very low. |
| − | + | ||
Metabolic engineering refers to the alteration of genetic and/or regulatory circuitry within organisms. The native circuitry involves a series of enzymes that perform biochemical reactions that function to convert raw substrates into products that are required for the organism’s survival. When this native circuitry is altered using metabolic engineering techniques, the goal is to use the host organisms as machines that produce valuable materials in large quantities and at low cost. | Metabolic engineering refers to the alteration of genetic and/or regulatory circuitry within organisms. The native circuitry involves a series of enzymes that perform biochemical reactions that function to convert raw substrates into products that are required for the organism’s survival. When this native circuitry is altered using metabolic engineering techniques, the goal is to use the host organisms as machines that produce valuable materials in large quantities and at low cost. | ||
| − | Common strategies employed in metabolic engineering are: | + | [[Image:Fa16 M2D2 pathway example.png|thumb|300px|right|'''Example illustrating pathway to desired product.''']]Common strategies employed in metabolic engineering are: |
*Increasing expression of a gene that encodes an enzyme responsible for a rate-limiting step. | *Increasing expression of a gene that encodes an enzyme responsible for a rate-limiting step. | ||
*Inhibiting competing pathways that divert substrate away from the pathway of interest. | *Inhibiting competing pathways that divert substrate away from the pathway of interest. | ||
| Line 15: | Line 14: | ||
*Altering the structure of the enzyme such that yield is increased. | *Altering the structure of the enzyme such that yield is increased. | ||
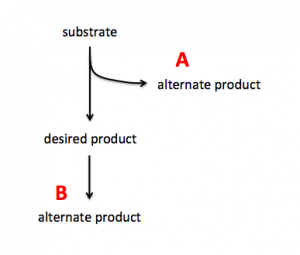
| − | + | To increase production of a desired product, you can either target proteins that use the substrate to generate alternate products (see A in image on right) or you can target proteins that use the desired product to generate alternate products (see B in image on right). In A the substrate is siphoned away from the reaction that generates the desired product and in B the desired product is used as substrate in a subsequent reaction. By eliminating the proteins that catalyze the reactions that result in alternate products, you can potentially increase production of your desired product. | |
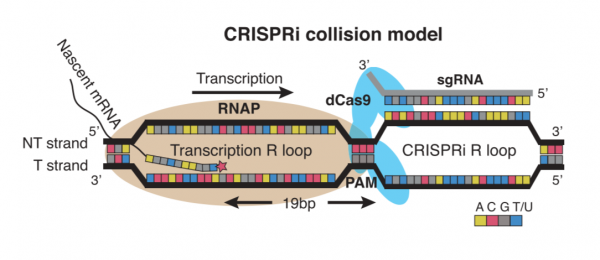
| − | To manipulate the metabolism of ''E. coli'', | + | To manipulate the metabolism of ''E. coli'', the CRISPRi system was used. Unlike more commonly used CRISPR-based technologies, CRISPRi is used to modulate expression from the genome rather than to modify the genome. This distinction is due to the use of an enzymatically-inactive dCas9 (or ‘dead’ Cas9) protein. Because dCas9 is enzymatically inactive, it is unable to cleave the DNA upon binding to the targeted sequence in the host genome. The lack of DNA cleavage results in gene silencing through impeding RNA polymerase binding, transcription factor binding, and/or transcription elongation. This method of repression referred to as CRISPRi collision and depicted in the schematic below. |
| + | |||
| + | [[Image:Fa20 M3D1 CRISPRi collision model.png|thumb|600px|center|]] | ||
==Protocols== | ==Protocols== | ||
| − | ===Part | + | The focus for today is to understand the key concepts and methods that were used to generate the data you will critically evaluate in this module. First, it is important to consider how targets were selected for the metabolic engineering approach. Second, you will review the CRISPRi technology used for the metabolic engineering approach. |
| − | + | ||
| + | ===Part 1: Review ''E. coli'' anaerobic fermentative metabolism=== | ||
| + | |||
| + | Across many fields of science, ''E. coli'' is a valuable tool in research related to genetic systems, metabolic pathways, and physiological responses. It is often called the 'workhorse' of bench science. There are many reasons for this role including: 1) ''E. coli'' cultures are easy to grow in the laboratory, 2) ''E. coli'' is a genetically tractable system, 3) ''E. coli'' metabolic pathways are defined and, 4) the breadth of research available for ''E. coli'' in literature. For all of these reasons, ''E. coli'' is ideal for use this module! | ||
| + | |||
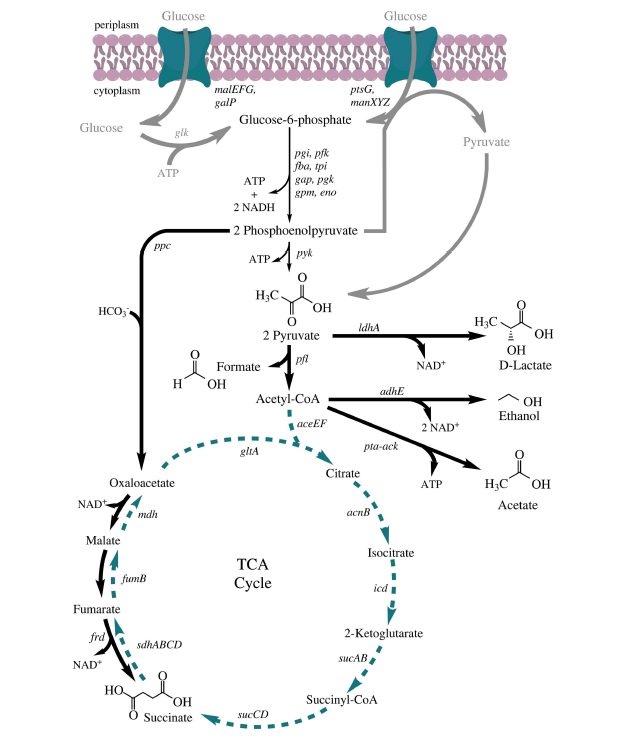
| + | We will take advantage of the genetic tools available for engineering the genetics of ''E. coli'' to increase ethanol yield. In ''E. coli'', ethanol is produced via the mixed-acid pathway which is used by cells under fermentative conditions to maintain redox balance. With your laboratory partner, review the metabolic pathway map below. Specifically, identify at which step ethanol is produced. | ||
| + | |||
| + | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | ||
| + | *Which gene(s) are responsible for ethanol production? Provide a brief description of the reaction (ie what is substrate? are biproducts generated?). | ||
| + | *Which gene(s) might you target using the CRISPRi system to increase the availability of substrate for ethanol production? | ||
| + | *Which gene(s) might you target using the CRISPRi system to decrease the amount of substrate used to generate products in steps downstream of ethanol production? | ||
| + | *From the list of possible gene targets, which do you think is the most promising candidate? Why? | ||
| + | |||
| + | [[Image:Fa16 M2D2 fermentation pathway.png|thumb|700px|center|'''Mixed-acid fermentation in ''E. coli'''''. Image from [[Media:MetEngEcoli review.pdf |Metabolic engineering of ''Escherichia coli'' for production of mixed-acid fermentation end products.]] ''Frontiers in Bioengineering and Biotechnology.'' (2014)2:1-12. ]]<br> | ||
| − | + | ===Part 2: Research CRISPRi system=== | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | Review and discuss the Introduction and the first Results sub-section ('A minimal CRISPRi system consists of a single protein and RNA and can effectively silence transcription initiation and elongation') from the following journal article with your laboratory partner: | |
| − | + | Lei ''et al.'' "[[Media:Fa20 M3D1 CRISPRi reference.pdf |Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression.]]" ''Cell''. (2013) 152:1173-1183. | |
| − | + | In this article, the researchers describe a modified CRISPR system, referred to as CRISPR interference (CRISPRi), that inhibits transcription of genes that are targeted using single guide RNA (sgRNA) molecules. The goal of this exercise is to understand how the sgRNA molecules are used to target specific genes. In the CRISPRi system, sgRNA forms a complex with dCas9 and this complex binds to the sequence in the genome that is complementary to the sgRNA molecule. Thus, the sgRNA targets a specific gene sequence in the host genome. When the sgRNA / dCas9 complex binds, transcription of the gene is inhibited. | |
| − | + | Transcription can be blocked by targeting sgRNA molecules to the promoter or coding region of a specific gene. The promoter is the region upstream of the start codon and contains the RNAP binding site, or the -10 and -35. When the sgRNA / dCas9 complex binds to the promoter, transcription initiation is inhibited because RNAP is unable to bind to the promoter. The coding region refers to the gene sequence. When the sgRNA is targeted to the coding region, transcription elongation is stalled because RNAP is unable to traverse beyond the location of the sgRNA / dCas9 complex. | |
| − | + | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following with your partner: | |
| + | *Briefly, describe how the CRISPRi system is different than the native CRISPR system that was discussed during lecture. | ||
| + | **Hint: Which two components were modified? How? | ||
| + | *In the first paragraph of the Results sub-section, what are the researchers testing? | ||
| + | *Describe the red fluorescent protein (mRFP)-based reporter system. | ||
| + | **Where was the mRFP gene sequence inserted? | ||
| + | **What does this system enable the researchers to test? | ||
| + | *For Fig. 2C, describe the schematic of the mRFP-based reporter system (top of the panel). | ||
| + | **What are each of the components and what is the purpose of each? | ||
| + | **What does it mean if RFP is expressed from this construct? If RFP is not expressed from this construct? | ||
| + | *For Fig. 2C, explain the data that are shown (bottom of the panel). | ||
| + | **What is the control? | ||
| + | **What do these data tell you about how to best target the coding region of a gene using sgRNA molecules and the CRISPRi system? | ||
| + | *For Fig. 2D, describe the schematic of the experimental approach (top of panel). | ||
| + | **How does this experiment relate to what is shown in Fig. 2C? | ||
| + | **The output from this experimental approach is RFP. Where is the RFP insert? How does this approach relate to the mRFP-based reporter system? | ||
| + | *For Fig. 2D, explain the data that are shown (bottom of the panel). | ||
| + | **What is the control? | ||
| + | **What do these data tell you about how to best target the promoter region of a gene using sgRNA molecules and the CRISPRi system? | ||
| + | *Lastly, given the results from this article what is the best approach for targeting the promising candidate you identified above in Part 1? | ||
| + | **Should you design an sgRNA that targets, or binds, the promoter or the coding region? The template or the non-template strand? | ||
| + | **Where in the promoter or coding region should the sgRNA bind (ie the -35 or the middle of the gene)? | ||
| − | + | ==Navigation links== | |
| + | Next day: [[20.109(F20):M3D2 | Align gRNA sequences with genomic targets]]<br> | ||
| + | Previous day: [[20.109(F20):M2D7 | Examine putative small molecule binders for common features]] ?? | ||
Latest revision as of 18:34, 22 July 2020
Contents
Introduction
In this module you will build upon the progress of former 109ers! In previous semesters, students engineered the the metabolism of E. coli in an attempt to increase the production of a commercially relevant product. Specifically, metabolic engineering was used to increase the ethanol yield via manipulating the fermentation pathway native to E. coli cells. Ethanol, or ethyl alcohol, is used as fuel – most commonly as a biofuel additive in gasoline. Current methods for producing ethanol involve the use of agricultural feedstocks and concern is mounting given the effect this may have on food prices and resources. Using E. coli as an ethanol-generating machine is promising; however, the ethanol output of a native cell is very low.
Metabolic engineering refers to the alteration of genetic and/or regulatory circuitry within organisms. The native circuitry involves a series of enzymes that perform biochemical reactions that function to convert raw substrates into products that are required for the organism’s survival. When this native circuitry is altered using metabolic engineering techniques, the goal is to use the host organisms as machines that produce valuable materials in large quantities and at low cost.
Common strategies employed in metabolic engineering are:- Increasing expression of a gene that encodes an enzyme responsible for a rate-limiting step.
- Inhibiting competing pathways that divert substrate away from the pathway of interest.
- Incorporating genes from other organisms into the host.
- Altering the structure of the enzyme such that yield is increased.
To increase production of a desired product, you can either target proteins that use the substrate to generate alternate products (see A in image on right) or you can target proteins that use the desired product to generate alternate products (see B in image on right). In A the substrate is siphoned away from the reaction that generates the desired product and in B the desired product is used as substrate in a subsequent reaction. By eliminating the proteins that catalyze the reactions that result in alternate products, you can potentially increase production of your desired product.
To manipulate the metabolism of E. coli, the CRISPRi system was used. Unlike more commonly used CRISPR-based technologies, CRISPRi is used to modulate expression from the genome rather than to modify the genome. This distinction is due to the use of an enzymatically-inactive dCas9 (or ‘dead’ Cas9) protein. Because dCas9 is enzymatically inactive, it is unable to cleave the DNA upon binding to the targeted sequence in the host genome. The lack of DNA cleavage results in gene silencing through impeding RNA polymerase binding, transcription factor binding, and/or transcription elongation. This method of repression referred to as CRISPRi collision and depicted in the schematic below.
Protocols
The focus for today is to understand the key concepts and methods that were used to generate the data you will critically evaluate in this module. First, it is important to consider how targets were selected for the metabolic engineering approach. Second, you will review the CRISPRi technology used for the metabolic engineering approach.
Part 1: Review E. coli anaerobic fermentative metabolism
Across many fields of science, E. coli is a valuable tool in research related to genetic systems, metabolic pathways, and physiological responses. It is often called the 'workhorse' of bench science. There are many reasons for this role including: 1) E. coli cultures are easy to grow in the laboratory, 2) E. coli is a genetically tractable system, 3) E. coli metabolic pathways are defined and, 4) the breadth of research available for E. coli in literature. For all of these reasons, E. coli is ideal for use this module!
We will take advantage of the genetic tools available for engineering the genetics of E. coli to increase ethanol yield. In E. coli, ethanol is produced via the mixed-acid pathway which is used by cells under fermentative conditions to maintain redox balance. With your laboratory partner, review the metabolic pathway map below. Specifically, identify at which step ethanol is produced.
In your laboratory notebook, complete the following:
- Which gene(s) are responsible for ethanol production? Provide a brief description of the reaction (ie what is substrate? are biproducts generated?).
- Which gene(s) might you target using the CRISPRi system to increase the availability of substrate for ethanol production?
- Which gene(s) might you target using the CRISPRi system to decrease the amount of substrate used to generate products in steps downstream of ethanol production?
- From the list of possible gene targets, which do you think is the most promising candidate? Why?
Part 2: Research CRISPRi system
Review and discuss the Introduction and the first Results sub-section ('A minimal CRISPRi system consists of a single protein and RNA and can effectively silence transcription initiation and elongation') from the following journal article with your laboratory partner:
Lei et al. "Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression." Cell. (2013) 152:1173-1183.
In this article, the researchers describe a modified CRISPR system, referred to as CRISPR interference (CRISPRi), that inhibits transcription of genes that are targeted using single guide RNA (sgRNA) molecules. The goal of this exercise is to understand how the sgRNA molecules are used to target specific genes. In the CRISPRi system, sgRNA forms a complex with dCas9 and this complex binds to the sequence in the genome that is complementary to the sgRNA molecule. Thus, the sgRNA targets a specific gene sequence in the host genome. When the sgRNA / dCas9 complex binds, transcription of the gene is inhibited.
Transcription can be blocked by targeting sgRNA molecules to the promoter or coding region of a specific gene. The promoter is the region upstream of the start codon and contains the RNAP binding site, or the -10 and -35. When the sgRNA / dCas9 complex binds to the promoter, transcription initiation is inhibited because RNAP is unable to bind to the promoter. The coding region refers to the gene sequence. When the sgRNA is targeted to the coding region, transcription elongation is stalled because RNAP is unable to traverse beyond the location of the sgRNA / dCas9 complex.
In your laboratory notebook, complete the following with your partner:
- Briefly, describe how the CRISPRi system is different than the native CRISPR system that was discussed during lecture.
- Hint: Which two components were modified? How?
- In the first paragraph of the Results sub-section, what are the researchers testing?
- Describe the red fluorescent protein (mRFP)-based reporter system.
- Where was the mRFP gene sequence inserted?
- What does this system enable the researchers to test?
- For Fig. 2C, describe the schematic of the mRFP-based reporter system (top of the panel).
- What are each of the components and what is the purpose of each?
- What does it mean if RFP is expressed from this construct? If RFP is not expressed from this construct?
- For Fig. 2C, explain the data that are shown (bottom of the panel).
- What is the control?
- What do these data tell you about how to best target the coding region of a gene using sgRNA molecules and the CRISPRi system?
- For Fig. 2D, describe the schematic of the experimental approach (top of panel).
- How does this experiment relate to what is shown in Fig. 2C?
- The output from this experimental approach is RFP. Where is the RFP insert? How does this approach relate to the mRFP-based reporter system?
- For Fig. 2D, explain the data that are shown (bottom of the panel).
- What is the control?
- What do these data tell you about how to best target the promoter region of a gene using sgRNA molecules and the CRISPRi system?
- Lastly, given the results from this article what is the best approach for targeting the promising candidate you identified above in Part 1?
- Should you design an sgRNA that targets, or binds, the promoter or the coding region? The template or the non-template strand?
- Where in the promoter or coding region should the sgRNA bind (ie the -35 or the middle of the gene)?
Next day: Align gRNA sequences with genomic targets