Difference between revisions of "20.109(F20):M1D3"
Noreen Lyell (Talk | contribs) (→Part 1: Begin antibody staining for γH2AX assay) |
Noreen Lyell (Talk | contribs) (→Part 1: Begin antibody staining for γH2AX assay) |
||
| (28 intermediate revisions by 2 users not shown) | |||
| Line 16: | Line 16: | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| − | In your experiment, you will use a primary antibody to bind the γH2AX foci. Then a secondary antibody will be used that is specific to the conserved region of the primary antibody. The use of secondary antibodies allows researchers to tag the primary antibody. In our assay, the tag is a 488 nm fluorescent dye that will enable us to visualize double-strand breaks via microscopy. As a reminder, during the last laboratory session MCL-5 cells were treated with H<sub>2</sub>O<sub>2</sub> +/- As for the H2AX assay. Today we will discuss how to permeabilize the cells, which will enable the antibodies to enter the cells and bind γH2AX. | + | In your experiment, you will use a primary antibody to bind the γH2AX foci. Then a secondary antibody will be used that is specific to the conserved region of the primary antibody. The use of secondary antibodies allows researchers to tag the primary antibody. In our assay, the tag is a 488 nm fluorescent dye that will enable us to visualize double-strand breaks via microscopy. As a reminder, during the last laboratory session MCL-5 cells were treated with H<sub>2</sub>O<sub>2</sub> +/- As for the γH2AX assay. Today we will discuss how to permeabilize the cells, which will enable the antibodies to enter the cells and bind γH2AX. |
==Protocols== | ==Protocols== | ||
| Line 22: | Line 22: | ||
===Part 1: Begin antibody staining for γH2AX assay=== | ===Part 1: Begin antibody staining for γH2AX assay=== | ||
| − | <font color = #0d368e>'''To ensure the steps included below are clear, please watch the video tutorial | + | <font color = #0d368e>'''To ensure the steps included below are clear, please watch the video tutorial linked here: [[https://www.dropbox.com/s/znamgumq97928ys/H2AX.mp4?dl=0 H2AX Staining]]. The steps are detailed below so you can follow along!'''</font color> |
[[Image:IMG_5503.JPG|thumb|right|350px|'''Immunofluorescence staining chamber''']] | [[Image:IMG_5503.JPG|thumb|right|350px|'''Immunofluorescence staining chamber''']] | ||
| Line 43: | Line 43: | ||
#Aspirate the block solution and add 150 μL of the diluted primary antibody solution to each coverslip before moving the next. Do not let the coverslips dry! | #Aspirate the block solution and add 150 μL of the diluted primary antibody solution to each coverslip before moving the next. Do not let the coverslips dry! | ||
#Carefully move your staining chambers to the 4 °C cooler. | #Carefully move your staining chambers to the 4 °C cooler. | ||
| − | + | #Incubate samples at 4 °C in the primary antibody solution for ~48 h. | |
| + | #Wash 3x with TBS-Triton | ||
| + | #Replace the primary antibody solution with the secondary antibody solution, Alexa Fluor 488 goat anti-mouse diluted 1:200 in blocking solution 1 h prior to imaging. | ||
| − | ===Part 2: | + | ===Part 2: Participate in group paper discussion=== |
| + | We will finish today with a discussion concerning the following research article: | ||
| − | + | Weingeist, D. M., et al. "[[Media:Weingest 2013 Single cell microarray enables high throughput evaluation of DNA double strand breaks and DNA repair inhibitors.pdf|Single-cell microarray enables high-throughput evaulation of DNA double-strand breaks and DNA repair inhibitors.]]" ''Cell Cycle''. (2013) 126:907-915. | |
| − | + | '''From the Introduction''' | |
| − | + | Consider the key components of an introduction: | |
| + | *What is the big picture? | ||
| + | *Is the importance of this research clear? | ||
| + | *Are you provided with the information you need to understand the research? | ||
| + | *Do the authors include a preview of the key results? | ||
| − | ''' | + | '''From the Results''' |
| − | + | ||
| − | + | Carefully examine the figures. First, read the captions and use the information to 'interpret' the data presented within the image. Second, read the text within the results section that describes the figure. | |
| − | + | *Do you agree with the conclusion(s) reached by the authors? | |
| − | + | *What controls were included and are they appropriate for the experiment performed? | |
| − | + | *Are you convinced that the data are accurate and/or representative? | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | '''From the Discussion''' | |
| − | + | Consider the following components of a discussion: | |
| − | + | *Are the results summarized? | |
| − | + | *Did the authors 'tie' the data together into a cohesive and well-interpreted story? | |
| − | + | *Do the authors overreach when interpreting the data? | |
| − | + | *Are the data linked back to the big picture from the introduction? | |
| − | + | ||
| − | + | <font color = #4a9152 >'''In your laboratory notebook,'''</font color> complete the following: | |
| − | + | *Based on your reading and the group discussion of the article, answer the questions above. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | # | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
==Reagents list== | ==Reagents list== | ||
| − | |||
*permeabilization buffer: 0.2% Triton in Tris buffer saline (TBS) (from Invitrogen) | *permeabilization buffer: 0.2% Triton in Tris buffer saline (TBS) (from Invitrogen) | ||
| − | *blocking buffer: 1% bovine serum albumin (BSA) in TBS (from Sigma) | + | *blocking buffer: 1% bovine serum albumin (BSA) in TBS (BSA from Sigma) |
*1:1000 primary antibody to γH2AX, mouse (from Millipore) | *1:1000 primary antibody to γH2AX, mouse (from Millipore) | ||
| − | *1:200 Alexa Fluor | + | *1:200 Alexa Fluor 488 goat anti-mouse IgG (from ThermoFisher) |
| − | * | + | *1:1000 DAPI (from ThermoFisher) |
| − | + | *Fluoromount G (from Southern Biotech) | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | * | + | |
| − | + | ||
==Navigation links== | ==Navigation links== | ||
| − | Next day: | + | Next day: [[20.109(F20):M1D4 | Image repair foci experiment and quantify results]]<br> |
| + | Previous day: [[20.109(F20):M1D2 | Prepare and treat cells for repair foci experiment]]<br> | ||
Latest revision as of 23:14, 10 September 2020
Contents
Introduction
You will use commercially available antibodies to identify γH2AX foci in your experiment. The ability to bind specific proteins using antibodies, or immunoglobulins, is critical in immuno-fluorescence labeling analysis. Antibodies are typically 'raised' in mammalian hosts. Most commonly mice, rabbits, and goats are used, but antibodies can also be raised in sheep, chickens, rats, and even humans. The protein used to raise an antibody is called the antigen and the portion of the antigen that is recognized by an antibody is called the epitope. Some antibodies are monoclonal, or more appropriately “monospecific,” and recognize one epitope, while other antibodies, called polyclonal antibodies, are in fact antibody pools that recognize multiple epitopes. Antibodies can be raised not only to detect specific amino acid sequences, but also post-translational modifications and/or secondary structure. Therefore, antibodies can be used to distinguish between modified (for example, phosphorylated or glycoslyated proteins) and unmodified protein.
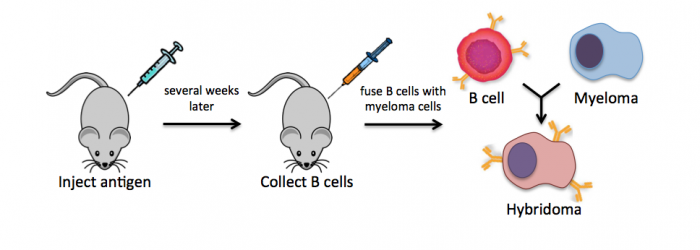
Monoclonal antibodies overcome many limitations of polyclonal pools in that they are specific to a particular epitope and can be produced in unlimited quantities. However, more time is required to establish these antibody-producing cells, called hybridomas, and it is a more expensive endeavor. In this process, normal antibody-producing B cells are fused with immortalized B cells, derived from myelomas, by chemical treatment with a limited efficiency. To select only heterogeneously fused cells, the cultures are maintained in medium in which myeloma cells alone cannot survive (often HAT medium). Normal B cells will naturally die over time with no intervention, so ultimately only the fused cells, called hybridomas, remain. A fused cell with two nuclei can be resolved into a stable cell line after mitosis.
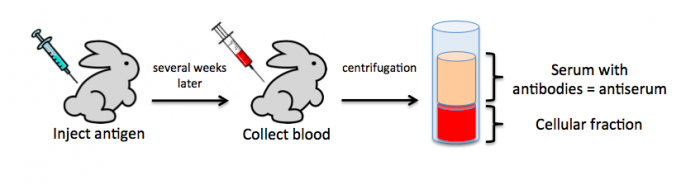
To raise polyclonal antibodies, the antigen of interest is first purified and then injected into an animal. To elicit and enhance the animal’s immunogenic response, the antigen is often injected multiple times over several weeks in the presence of an immune-boosting compound called adjuvant. After some time, usually 4 to 8 weeks, samples of the animal’s blood are collected and the cellular fraction is removed by centrifugation. What is left, called the serum, can then be tested in the lab for the presence of specific antibodies. Even the very best antisera have no more than 10% of their antibodies directed against a particular antigen. The quality of any antiserum is judged by the purity (that it has few other antibodies), the specificity (that it recognizes the antigen and not other spurious proteins) and the concentration (sometimes called titer). Animals with strong responses to an antigen can be boosted with the antigen and then bled many times, so large volumes of antisera can be produced. However animals have limited life-spans and even the largest volumes of antiserum will eventually run out, requiring a new animal. The purity, specificity and titer of the new antiserum will likely differ from those of the first batch. High titer antisera against bacterial and viral proteins can be particularly precious since these antibodies are difficult to raise; most animals have seen these immunogens before and therefore don’t mount a major immune response when immunized. Antibodies against toxic proteins are also challenging to produce if they make the animals sick.
In your experiment, you will use a primary antibody to bind the γH2AX foci. Then a secondary antibody will be used that is specific to the conserved region of the primary antibody. The use of secondary antibodies allows researchers to tag the primary antibody. In our assay, the tag is a 488 nm fluorescent dye that will enable us to visualize double-strand breaks via microscopy. As a reminder, during the last laboratory session MCL-5 cells were treated with H2O2 +/- As for the γH2AX assay. Today we will discuss how to permeabilize the cells, which will enable the antibodies to enter the cells and bind γH2AX.
Protocols
Part 1: Begin antibody staining for γH2AX assay
To ensure the steps included below are clear, please watch the video tutorial linked here: [H2AX Staining]. The steps are detailed below so you can follow along!
- Obtain your 12-well plates from the front laboratory bench.
- Gather an aliquot of 1 X TBS from the front laboratory bench.
- Prepare 1.2 mL solution of 0.2% Triton X-100 (v/v) in 1X TBS in a micro centrifuge tube. 10% Triton stock is at the front laboratory bench.
- Prepare 2.5 mL solution of 1% BSA (v/v) in 1X TBS in 15ml conical tube. 10% BSA stock is at the front bench.
- One of the preparations will be the blocking solution used in Step #8 and the other preparation will be used in Step #9 for the primary antibody solution.
- Obtain a staining chamber from the front bench and add a damp paper towel to each side of the parafilm. Label parafilm with experimental details.
- Obtain a fine gauge (26 3/8) needle and a pair of tweezers from the front laboratory bench.
- Carefully press the tip of the needle against the benchtop to bend it into a right angle such that the beveled side of the needle is the interior angle.
- Use the 'hook' created with the needle to lift the coverslip from the bottom of the well, then use the tweezers to 'catch' the coverslip.
- Practice plates with coverslips will be available at the front laboratory bench.
- When you are confident with your ability to retrieve the coverslips from the wells, move one coverslip from each condition from your 12-well plates to the staining chamber. Cell-side UP!
- The cell-side of the coverslip is the side that was facing up in the well of the 12-well plate.
- Quickly permeabilize the cells by adding 150 μL of the 0.2% Triton X-100/TBS solution to each coverslip and incubate for 10 min at room temperature.
- Aspirate the 0.2% Triton X-100/TBS solution and add 150 μL of BSA blocking solution to each coverslip, then incubate for 60 min at room temperature.
- With 15 min remaining of the blocking solution incubation, prepare the primary antibody.
- Dilute the mouse anti-γH2AX antibody 1:1000 in the 1.2 mL aliquot of BSA blocking solution.
- Aspirate the block solution and add 150 μL of the diluted primary antibody solution to each coverslip before moving the next. Do not let the coverslips dry!
- Carefully move your staining chambers to the 4 °C cooler.
- Incubate samples at 4 °C in the primary antibody solution for ~48 h.
- Wash 3x with TBS-Triton
- Replace the primary antibody solution with the secondary antibody solution, Alexa Fluor 488 goat anti-mouse diluted 1:200 in blocking solution 1 h prior to imaging.
Part 2: Participate in group paper discussion
We will finish today with a discussion concerning the following research article:
Weingeist, D. M., et al. "Single-cell microarray enables high-throughput evaulation of DNA double-strand breaks and DNA repair inhibitors." Cell Cycle. (2013) 126:907-915.
From the Introduction
Consider the key components of an introduction:
- What is the big picture?
- Is the importance of this research clear?
- Are you provided with the information you need to understand the research?
- Do the authors include a preview of the key results?
From the Results
Carefully examine the figures. First, read the captions and use the information to 'interpret' the data presented within the image. Second, read the text within the results section that describes the figure.
- Do you agree with the conclusion(s) reached by the authors?
- What controls were included and are they appropriate for the experiment performed?
- Are you convinced that the data are accurate and/or representative?
From the Discussion
Consider the following components of a discussion:
- Are the results summarized?
- Did the authors 'tie' the data together into a cohesive and well-interpreted story?
- Do the authors overreach when interpreting the data?
- Are the data linked back to the big picture from the introduction?
In your laboratory notebook, complete the following:
- Based on your reading and the group discussion of the article, answer the questions above.
Reagents list
- permeabilization buffer: 0.2% Triton in Tris buffer saline (TBS) (from Invitrogen)
- blocking buffer: 1% bovine serum albumin (BSA) in TBS (BSA from Sigma)
- 1:1000 primary antibody to γH2AX, mouse (from Millipore)
- 1:200 Alexa Fluor 488 goat anti-mouse IgG (from ThermoFisher)
- 1:1000 DAPI (from ThermoFisher)
- Fluoromount G (from Southern Biotech)
Next day: Image repair foci experiment and quantify results