Difference between revisions of "20.109(F20):M1D2"
Noreen Lyell (Talk | contribs) (→Introduction) |
Noreen Lyell (Talk | contribs) (→Introduction) |
||
| Line 19: | Line 19: | ||
As mentioned above, the final step in BER is a ligation reaction that seals the helix. Arsenite has been shown to enhance the genotoxicity of several mutagens and inhibit ligase activity. Thus, the experiments in this module are designed to test the combined effects of exposure to MMS, a DNA damaging agent, and As, a chemical known to decrease DNA repair capacity by inhibiting strand ligation. | As mentioned above, the final step in BER is a ligation reaction that seals the helix. Arsenite has been shown to enhance the genotoxicity of several mutagens and inhibit ligase activity. Thus, the experiments in this module are designed to test the combined effects of exposure to MMS, a DNA damaging agent, and As, a chemical known to decrease DNA repair capacity by inhibiting strand ligation. | ||
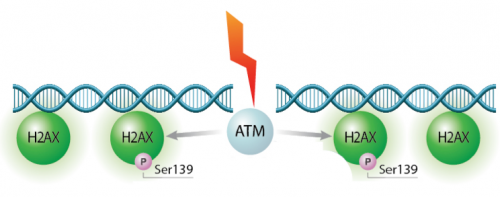
| − | We will test the hypothesis for this module using two experiments: the γH2AX assay and the CometChip assay. Today we will start the γH2AX experiment. In eukaryotes, including humans, DNA is tightly wound around histone groups. H2AX is a member of the core group of histones that contributes to nucleosome formation and DNA structure. When a DNA double-strand break is introduced into the genome, the H2AX histones near the break are phosphorylated by the ATM kinase at residue Ser-139. Upon phosphorylation H2AX is referred to as gamma-H2AX. Given that only H2AX histones near the site of DNA damage are phosphorylated, γH2AX is a useful target when determining the abundance and location of double-strand breaks. | + | We will test the hypothesis for this module using two experiments: the γH2AX assay and the CometChip assay. Today we will start the γH2AX experiment. In eukaryotes, including humans, DNA is tightly wound around histone groups. H2AX is a member of the core group of histones that contributes to nucleosome formation and DNA structure. When a DNA double-strand break is introduced into the genome, the H2AX histones near the break are phosphorylated by the ATM kinase at residue Ser-139. Upon phosphorylation H2AX is referred to as gamma-H2AX. Given that only H2AX histones near the site of DNA damage are phosphorylated, γH2AX is a useful target when determining the abundance and location of double-strand breaks. It is important to highlight that the DNA damage expected to occur in response to H<sub>2</sub>O<sub>2</sub> treatment is single-stranded breaks. So why are we using the γH2AX assay to measure double-stranded breaks? When DNA is damaged by multiple single-stranded breaks, double-stranded breaks can occur. |
[[Image:Fa16 M1D5 H2AX-P.png|thumb|center|500px|'''H2AX is phosphorylated in response to DNA double-strand breaks.''']] | [[Image:Fa16 M1D5 H2AX-P.png|thumb|center|500px|'''H2AX is phosphorylated in response to DNA double-strand breaks.''']] | ||
Revision as of 00:48, 16 July 2020
Contents
Introduction
The goal of this module is to test the hypothesis that As leads to higher levels of H2O2-induced DNA damage in an effort to address potential public health risks associated with combined exposures to hazardous compounds. Before we discuss the specific experiments, let's review the important background information.
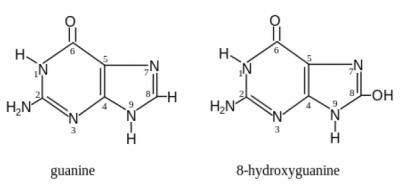
Hydrogen peroxide (H2O2) is an oxidizing agentNormal cell tissues have a basal level of DNA damage due to cell processes involved in cellular metabolism. For example, electrons can escape the electron transport chain and result in the formation of superoxide. Furthermore, defense mechanisms employed to protect the host from bacterial infection involved the release of reactive oxygen species. These reactive oxygen species are implicated in causing more than 20 types of DNA base lesions. One of the most common types of damage is the change of guanine to 8-hydroxyguanine.
Base excision repair (BER) pathway repairs damaged bases
The Base Excision Repair (BER) pathway corrects DNA damage, specifically the removal of small, non-helix distorting lesions in DNA caused by damaged bases. These lesions often result from oxidation, alkylation, deamination, and depuriniation/depryrimidination. If base lesions are not repaired, non-canonical base pairing can occur, which may result in the incorporation of an incorrect base during replication. To prevent mutations and maintain integrity of the genome, the BER pathway evolved as a highly conserved repair mechanism in both E. coli and mammals. Thus, this pathway is responsible for repairing damage before a mutation results.
The core BER pathway includes only four proteins that function to remove the damaged base and replace it with the correct base. First, a DNA glycosylase recognizes that a damaged base is present in the DNA and cleaves an N-glycosidic bond, which creates an apurinic or apyrimidinic site (referred to as an AP site in both cases). Different DNA glycosylases recognize different types of base lesions. Second, the DNA backbone is cleaved to create a single-strand DNA nick by either a DNA AP endonuclease or a DNA AP lyase. Next, a DNA polymerase incorporates the correct base using the sister strand as a template. Last, a DNA ligase completes the repair by sealing the single-strand nick, which restores integrity to the helix. For a more detailed description of the BER pathway, read this review by Robertson et al.
Arsenite (As) inhibits ligase activity
As mentioned above, the final step in BER is a ligation reaction that seals the helix. Arsenite has been shown to enhance the genotoxicity of several mutagens and inhibit ligase activity. Thus, the experiments in this module are designed to test the combined effects of exposure to MMS, a DNA damaging agent, and As, a chemical known to decrease DNA repair capacity by inhibiting strand ligation.
We will test the hypothesis for this module using two experiments: the γH2AX assay and the CometChip assay. Today we will start the γH2AX experiment. In eukaryotes, including humans, DNA is tightly wound around histone groups. H2AX is a member of the core group of histones that contributes to nucleosome formation and DNA structure. When a DNA double-strand break is introduced into the genome, the H2AX histones near the break are phosphorylated by the ATM kinase at residue Ser-139. Upon phosphorylation H2AX is referred to as gamma-H2AX. Given that only H2AX histones near the site of DNA damage are phosphorylated, γH2AX is a useful target when determining the abundance and location of double-strand breaks. It is important to highlight that the DNA damage expected to occur in response to H2O2 treatment is single-stranded breaks. So why are we using the γH2AX assay to measure double-stranded breaks? When DNA is damaged by multiple single-stranded breaks, double-stranded breaks can occur.
Protocols
Part 1: Seed cells for γH2AX assay
To ensure the steps required for seeding cells are clear, the Instructor will provide a live demonstration of this process.
In your laboratory notebook, complete the following:
- Provide a written overview / description of the the procedure used to seed cells for the γH2AX assay (from the live demonstration).
- Calculate the volume of cell suspension added to each well for the γH2AX assay.
- The cell suspension is 1 M cells / mL and 25,000 cells were added into each well.
- For how long will the seeded cells incubate prior to treatment with H2O2 +/- As?
Part 2: Treat cells for γH2AX assay
A plate seeded according to the procedure demonstrated in Part 1 was used to treat cells using the H2O2 +/- As conditions detailed below.
Before you read through the protocols for the H2O2 +/- As treatments, it is important to consider what conditions will be assessed in this experiment.
With your laboratory partner, make a list of all of the conditions that will be tested. Use the following descriptions of the variables that are included in this experiment to assist you.
- Two concentrations of H2O2 will be tested: 0 μM and 20 μM
- Three concentrations of As will be tested: 0 μM, 1 μM, and 10 μM
- Each concentration of H2O2 will be tested with each concentration of As.
- Four timepoints will be used for each condition tested: 0 min, 15 min, 30 min, and 60 min.
In your laboratory notebook, complete the following:
- Prepare a list or table of the conditions that will be used for the γH2AX assay.
- What conditions from your list or table are controls? For what does each condition control?
H2O2 treatment steps
Now that you know what conditions will be tested, the first step in the protocol is to treat the MCL-5 cells with H2O2.
- Retrieve the 12-well plate that was seeded with MCL-5 cells from the 37 °C incubator.
STEPS FOR ADHERING CELLS
- After aspirating off the media in the wells, add 1 mL of fresh media or media + 20 μM H2O2 to the appropriate wells.
- Incubate at 37 °C for 1 hour.
In your laboratory notebook, complete the following calculation:
- Calculate the dilution of H2O2 needed to have a final concentration of 20 μM.
- Stock concentration of H2O2 is 10 M.
- Volume of 20 μM H2O2 added to each well is 1 mL.
As treatment steps
- Retrieve the 12-well plate with the H2O2-treated MCL-5 cells from the incubator and use a pipette to remove the media from the wells.
- Add 1 mL of fresh media or media + As to the appropriate wells and incubate at 37 °C for the designated amount of time.
- Following treatment, transfer media to the As waste container and immediately add 400 μL of 4% paraformaldehyde to fix the cells.
- Incubate at room temperature for 10 min.
- Collect the 4% paraformaldehyde in the correct waste stream using a P1000 pipet.
- Wash with 500 μL of 1X PBS.
- Add 1X PBS then remove using a P1000 pipet. Collect the PBS in the correct waste stream.
- Complete a total of 2 times. Leaving 1 mL of 1X PBS on the cells in the final wash.
- Leave all wells with 1 mL PBS, parafilm the sides and move the fixed plates into the 4 °C cooler.
In your laboratory notebook, complete the following calculations:
- Calculate the dilutions of As needed to have final concentrations of 1 μM and 10 μM.
- Stock concentration of As is 100 mM.
- Volume of As solutions added to each well is 1 mL.
Reagents list
- 4% paraformaldehyde (from VWR)
- hydrogen peroxide (H2O2) (from Sigma)
- arsenite (As) (from Sigma)
- phosphate saline buffer (PBS) (from VWR)
Next day: Use immunoflourescence staining to assess repair foci experiment