20.109(S23):M2D5
Contents
Introduction
Before you can study the functional effect of the mutated Fet4 you generated in previous labs, you must make sure that the protein is expressed at the protein level. This is not a foregone conclusion as there are numerous examples of single point mutations which result in loss of protein expression. As a highly relevant example, in the Dix et. al paper you examined on M2D1. They found that their E406A point mutation in the Fet4 DNA sequence resulted in loss of protein expression compared to the wild type Fet4.
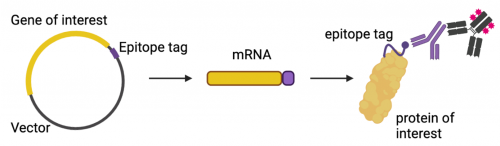
When working with understudied proteins, it is likely that commercial antibodies are not available to examine protein expression. Additionally, commercial antibodies can be expensive and may not be externally validated to guarantee their specificity. To avoid these issues, many recombinant proteins are made with peptide tags attached to the recombinant protein sequence at either the N-terminus or C-terminus. These peptide sequences can serve dual function. For example, in Mod1 you were able to use a 6x-His tag for affinity chromatography purification followed by using the same 6x-His tag to allow visualization of protein expression.
Other types of peptide tags are highly in identifying protein expression. Common tags are HA, FLAG, and V5. Each of these tags encodes a short peptide sequence that can be attached to the recombinant protein with minimal effects on native protein folding and activity. These tags allow for less expensive and more efficient study of recombinant proteins as high-affinity commercial antibodies can be produced and validated for widespread use.
The Fet4 protein you are studying has a C-terminal V5 tag incorporated into its cDNA. Not only does the tag allow you to study expression of a protein that lacks readily available commercial antibodies, the presence of the tag at the C-terminus allows you to confirm that the mutant Fet4 is expressed in its entirety, not truncated (shortened) through a frame shift or other unexpected mutation.
Protocols
Part 1: Induce expression of Fet4_mutant
For timing reasons, the induction steps were completed prior to class. So you understand how the cell suspensions you will use for the metal uptake assay were created, please review the steps below.
- Inoculated 5 mL of SD-R media with a colony of Alpha = W303α cells transformed with Fet4_mutant.
- Incubated the culture overnight at 30 °C with shaking at 220 rpm.
- Dilute the overnight culture 1:10 in 10 mL of fresh SD-R media.
- Incubate at 30 °C for 4 hours with shaking at 220 rpm.
- To induce Fet4_mutant protein expression, pellet cells and resuspend in 10ml of SD-G media.
- Incubate overnight at at 30 °C with shaking at 220 rpm.
In your laboratory notebook, answer the following questions:
- Why is the 4 hour incubation in fresh SD-R helpful to induction?
- Why do we need to change our media from SD-R to SD-G to induce yeast?
Part 2: Perform antibody staining to determine Fet4 transporter expression
Fix yeast and plate them on coverslips
- Obtain your 12-well plates with coated coverslips from the front laboratory bench.
- Obtain the following reagents from the front bench for your group:
- 1 culture of W303α yeast
- 1 culture of W303α yeast expressing Fet4
- 1 culture of W303α yeast expressing Fet4_mutant
- Media aliquot
- cuvettes
- Triturate your yeast culture to ensure even suspension, then transfer 1ml of each culture to a cuvette
- Add 1ml media to a fresh cuvette to use as a blank
- Determine the OD600 of each yeast culture
- Calculate the volume of each culture needed to make 2ml of each culture with an OD600 ~ 0.5 and dilute accordingly with fresh media in a fresh 15ml conical tube.
- Pellet cells by centrifugation at 900xg for 5 minutes and remove supernatent.
- Resuspend pellet in 1ml PFA, and incubate the cells at 30°C for 30 minutes.
- Pellet fixed cells at 900xg for 5 minutes and remove PFA.
- DO NOT aspirate PFA-- pipette it from the pellet and collect it for appropriate disposal.
- Wash pellet by resuspension in 1ml sorbitol-citrate buffer, then pellet cells by centrifugation at 900xg for 5 minutes.
- Aspirate the supernatent and resuspend the pellet in zymolyase.
- Prepare the zymolyase buffer by adding 30μL zymolyase (obtained at the front bench) to 3mL sorbitol-citrate buffer.
- Incubate the fixed yeast in 1 ml zymolase for 30 minutes at 30°C to digest the yeast cell wall.
- Pellet by spinning at 900g for 5 minutes
- Gather an aliquot of 1 X PBS from the front laboratory bench.
- Prepare 25 mL solution of 1% BSA (v/v) in 1X PBS in 50 mL conical tube. 10% BSA stock is at the front bench.
- One of the preparations will be the blocking solution used in Step #8 and the other preparation will be used in Step #9 for the primary antibody solution.
- Prepare 25 mL solution of 1% BSA (v/v) in 1X PBS in 50 mL conical tube. 10% BSA stock is at the front bench.
- Wash by resuspension 2x with 1 mL PBS + 1% BSA, aspirating in between each wash.
- Resuspend in 2 mls of PBS + 1% BSA
Complete primary staining steps
- Obtain your 12-well plates with coated coverslips from the front laboratory bench.
- Plate 1 ml of your yeast onto your coverslips. Allow to settle for 25 minutes.
- Prepare 1.2 mL solution of 0.1% Tween-20 (v/v) in 1X PBS in a micro centrifuge tube using the 10% Tween-20 stock is at the front laboratory bench.
- Obtain a staining chamber from the front bench and add a damp paper towel to each side of the parafilm.
- Label the parafilm with experimental details.
- Obtain a fine gauge (26 3/8) needle and a pair of tweezers from the front laboratory bench.
- Carefully press the tip of the needle against the benchtop to bend it into a right angle such that the beveled side of the needle is the interior angle.
- Use the 'hook' created with the needle to lift the coverslip from the bottom of the well, then use the tweezers to 'catch' the coverslip.
- Practice plates with coverslips will be available at the front laboratory bench.
- When you are confident with your ability to retrieve the coverslips from the wells, and your yeast have settled, move one coverslip from each condition from your 12-well plates to the staining chamber. Cell-side UP!
- The cell-side of the coverslip is the side that was facing up in the well of the 12-well plate.
- Quickly permeabilize the cells by adding 150 μL of the 0.1% Tween-20/PBS solution to each coverslip and incubate for 10 min at room temperature.
- Aspirate the 0.1% Tween-20/PBS solution and add 150 μL of BSA blocking solution to each coverslip, then incubate for 30 min at room temperature.
- With 15 min remaining of the blocking solution incubation, prepare the primary antibody.
- Dilute the mouse anti-V5 antibody 1:500 in the 1.2 mL aliquot of BSA blocking solution.
- Aspirate the block solution and add 150 μL of the diluted primary antibody solution to each coverslip before moving the next. Do not let the coverslips dry!
- Cover your staining chamber with the lid to incubate at room temperature.
- Incubate samples in the primary antibody solution for ~1 h.
Complete coverslip mounting steps
- Aspirate the primary antibody solution off the coverslip and add 150 μL of PBS per coverslip, let incubate at room temperature for 3 min covered, then aspirate.
- To add DAPI, dilute the DAPI stain 1:1000 in PBS and add 150 μL DAPI per coverslip.
- Incubate at room temperature for 5 min covered, then aspirate.
- Add PBS as in Step #2 for the final wash and leave for 3 min. Do not aspirate.
- Obtain glass slides from the front laboratory bench and label your slides with all of your experimental information and group name, add one drop (15 uL) of glycerol mounting media to the slide.
- Aspirate the final PBS wash and using tweezers place the coverslip cell-side down on the mounting media "spot" on the microscope slide. Try your best to avoid bubbles by slowly placing the coverslip over the mounting media.
- The cell-side of the coverslip is the side that was facing up in the staining chamber.
- Complete Steps #5-6 for coverslips from all of the coverslips you stained.
- Add one small drop of nail polish to each side of your coverslip to seal it to the glass slide.
In your laboratory notebook, complete the following:
- Why is zymolase used in this experiment? What would you predict occurs if this step is skipped?
- Why is it important to wash the antibody from the coverslip before imaging?
- What stain is used following the antibody? What cellular component is stained in this step? And why is this useful?
Reagents list
- Synthetic dropout - uracil (SD-U) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.192 % amino acid mix lacking uracil (Sigma), 2% glucose (BD Bacto), 0.1% adenine hemisulfate (Sigma)
- Synthetic dropout - raffinose (SD-R) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.192 % amino acid mix lacking uracil (Sigma), 2% raffinose (Sigma), 0.1% adenine hemisulfate (Sigma)
- Synthetic dropout - galactose (SD-G) media: 0.17% yeast nitrogen base without amino acid and ammonium sulfate (BD Bacto), 0.5% ammonium sulfate (Sigma), 0.192 % amino acid mix lacking uracil (Sigma), 2% raffinose (Sigma), 2% galactose (Sigma), 0.1% adenine hemisulfate (Sigma)
- 4% paraformaldehyde (Electron Microscopy Sciences)
- Sorbitol-citrate buffer: 1.2M sorbitol (Sigma), 10mM citric acid (Sigma), pH 7.0
- Zymolase (Zymo Research)
- permeabilization buffer: 0.1% Tween-20 in Phosphate buffer saline (PBS) (from Invitrogen)
- blocking buffer: 1% bovine serum albumin (BSA) in PBS (BSA from Sigma)
- 1:500 Alexafluor-488 conjugated primary antibody to V5 tag, mouse (from Genescript)
- 1:1000 DAPI (from ThermoFisher)
- 60% Glycerol (Sigma)
Next day: Analyze expression data and prepare metal uptake experiment