20.109(S23):M2D4
Contents
Introduction
In the previous laboratory session, you transformed your Fet4_mutant plasmid into yeast for further experimentation. While the transformed yeast are forming colonies, we will examine our sequencing results and prepare reagents for future experiments.To continue our experiments, we need to confirm that we achieved the correct point mutation, which you will do by aligning the results of your sanger sequencing experiment with our Fet4 template. However, since we are studying the resulting protein product, it is essential to confirm that your mutated protein expresses in our model system. There are different ways to accomplish this, but most techniques rely on the use of antibodies to visualize the expressed protein. Since your first step of the project involved manipulating Fet4, we are using a fusion tag to visualize our transporter, rather than an antibody raised against the protein itself. Additionally, due to schedule constraints, we will only prepare materials to examine mutated transporter expression today and complete the experiment on M2D6.
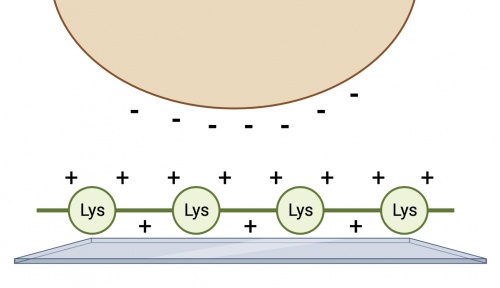
We have chosen to examine transporter expression using immunofluorescence rather than other approaches like Western blotting or flow cytometry. Partly, this approach was chosen to allow some qualitative assessment of protein expression within the cell as opposed to a yes or no answer. However, there is a challenge associated with this approach: yeast grow in suspension which makes microscopy challenging. To overcome this obstacle, we will coat our coverslips to encourage temporary adhesion of the yeast cells so that we can stain and image them. The coating we will use for this experiment is poly-D-lysine, a chemically synthesized amino acid chain with a net positive charge. This chain serves to facilitate cell adhesion by creating electrostatic interactions with the negatively charged yeast cell surface.
Protocols
Part 1: Participate in Communication Lab workshop
Our communication instructor, Dr. Chiara Ricci-Tam, will join us today for a discussion on oral presentations.
Part 2: Examine Fet4_mutant sequencing results
Your goal today is to analyze the sequencing data for you two potential mutant Fet4 clones - two independent colonies from your cloning reaction - and then decide which colony to proceed with to engineer yeast to become a cadmium sink.
Retrieve Fet4_mutant sequence results from Genewiz
- Your sequencing data is available from Genewiz. For easier access, the information was uploaded to the [20.109(S22):Class_data Class Data tab].
- Download the zip folder with your team sequencing results and confirm that there are 8 files saved in the folder.
- For each sequencing reaction, you should have one .ab1 file and one .seq file.
- Open one of the .ab1 files.
- This file contains the chromatogram for your sequencing reaction. Scroll through the sequence and ensure that the peaks are clearly defined and evenly spaced. Low signal (or peaks) or stacked peaks can provide incorrect base assignments in the sequence.
- Open one of the .seq files.
- This file contains the base assignments for your sequencing reaction. The bases are assigned by the software from the chromatogram sequence.
- The start of the a sequencing reaction result often contains several Ns, which indicates that the software was unable to assign a basepair.
In your laboratory notebook, complete the following:
- Given the chromatogram result, why might the software assign Ns in the start of the sequence?
- Visually inspect the chromatograms for all of your sequencing results.
- Do the peaks appear clearly defined or is there overlap? What might this indicate about the quality of your sequencing results?
- Do the peaks extend above the background signal? What might this indicate about the quality of your sequencing results?
Confirm point mutation in Fet4 sequence using SnapGene
You should align your sequencing data with a known sequence, in this case the Fet4 sequence from M2D1, to identify a successful mutation as well as any unintended base changes that may have occurred. There are several web-based programs for aligning sequences and still more programs that can be purchased. The steps for using SnapGene are below. Please feel free to use any program with which you are familiar.
- Generate a new DNA file that contains the Fet4 sequence you examined on M2D1.
- Generate an additional new DNA file that contains the results from the sequencing reaction completed by Genewiz.
- For each sequencing result you should generate a distinct new DNA file. Remember you should have a forward and reverse sequencing result for each of your clones!
- Paste the sequence text from your sequencing run into the new DNA file window. If there were ambiguous areas of your sequencing results, these will be listed as "N" rather than "A" "T" "G" or "C" and it's fine to include Ns in the query.
- The start and end of your sequencing may have several Ns. In this case it is best to omit these Ns by pasting only the 'good' sequence that is flanked by the ambiguous sequence.
- To confirm the mutation sequence in your clones, open one of the forward sequencing results files generated in the previous step.
- Select 'Tools' --> 'Align to Reference DNA Sequence...' --> 'Align Full Sequences...' from the toolbar.
- In the window, select the file that contains the sgRNA oligo sequence and click 'Open'.
- A new window will open with the alignment of the two sequences. The top line of sequence shows the results of the sequencing reaction and the bottom line shows the oligo you designed.
- Are there any discrepancies or differences between the two sequences? Scroll through the entire alignment to check the full sequencing result and note any basepair changes.
- Follow the above steps to examine all of your sequencing results. Remember: you used a forward and a reverse primer to interrogate both potential Fet4 point mutations.
- From the alignments, determine which Fet_mutant clone has the correct point mutation.
- If both clones contain the correct sequence choose either yeast transformation to use in the rest of your experiments. If only one is correct, then this is the transformant you will use. If neither of your plasmids carry the appropriate mutation, talk to your Instructor.
In your laboratory notebook, complete the following:
- Attach a screenshot for each alignment.
- Record which clone contains the correct point mutation.
Part 3: Prepare coverslips for immunofluorescence staining
While the yeast transformations are growing, you will prepare materials to confirm transporter expression in our yeast model system. As preparation for the IF experiment on M2D6, you will work in a sterile biosafety hood to coat coverslips for immunofluorescence staining.
- Collect a 12 well plate, a set of coverslips, and fine-tip forceps.
- These materials are packaged to maintain sterility and should only be opened in the hood.
- Remember to spray down all materials before placing them in the hood!
- Using the forceps, add coverslips to the 12 well plate according to the chart.
- Take an aliquot of sterile poly-D-lysine (PDL) into the hood.
- Add 1ml of PDL to each well that contains a coverslip.
- Allow coverslips to incubate with PDL for 20 minutes in the hood.
- Following incubation, aspirate the PDL and wash coverslips 2x with 1ml of DI water.
- Aspirate the final wash and allow the coverslips to dry in the hood for 20 minutes.
- Once coated coverslips have dried, label the plate with your team name/section and wrap the plate in parafilm for storage.
- Store plates at 4°C.
Reagents list
- Poly-D-Lysine 0.1mg/ml (Gibco)
Next day: Immunofluorescence staining